2CWI
 
 | | X-ray crystal structure analysis of recombinant wild-type canine milk lysozyme (apo-type) | | Descriptor: | Lysozyme C, milk isozyme, SULFATE ION | | Authors: | Akieda, D, Yasui, M, Aizawa, T, Yao, M, Watanabe, N, Tanaka, I, Demura, M, Kawano, K, Nitta, K. | | Deposit date: | 2005-06-20 | | Release date: | 2006-06-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Construction of an expression system of canine milk lysozyme in the methylotrophic yeast Pichia pastoris
To be Published
|
|
8YIE
 
 | | Crystal structure of GH13_30 alpha-glucosidase CmmB in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase | | Authors: | Saburi, W, Tagami, T, Yu, J, Ose, T, Yao, M, Mori, H. | | Deposit date: | 2024-02-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular mechanism for the substrate specificity of Arthrobacter globiformis M6 alpha-glucosidase CmmB, belonging to glycoside hydrolase family 13 subfamily 30
Food Biosci, 61, 2024
|
|
8YIF
 
 | | Crystal structure of GH13_30 alpha-glucosidase CmmB in complex with acarviosin | | Descriptor: | Acarviosin, Alpha-glucosidase | | Authors: | Saburi, W, Tagami, T, Yu, J, Ose, T, Yao, M, Mori, H. | | Deposit date: | 2024-02-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism for the substrate specificity of Arthrobacter globiformis M6 alpha-glucosidase CmmB, belonging to glycoside hydrolase family 13 subfamily 30
Food Biosci, 61, 2024
|
|
2E7D
 
 | | Crystal structure of a NEAT domain from Staphylococcus aureus | | Descriptor: | ACETATE ION, GLYCEROL, Hypothetical protein IsdH, ... | | Authors: | Suenaga, A, Tanaka, Y, Yao, M, Kumagai, I, Tanaka, I, Tsumoto, K. | | Deposit date: | 2007-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for multimeric heme complexation through a specific protein-heme interaction: the case of the third neat domain of IsdH from Staphylococcus aureus
J.Biol.Chem., 283, 2008
|
|
2DCU
 
 | | Crystal structure of translation initiation factor aIF2betagamma heterodimer with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Translation initiation factor 2 beta subunit, ... | | Authors: | Sokabe, M, Yao, M, Sakai, N, Toya, S, Tanaka, I. | | Deposit date: | 2006-01-16 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7C21
 
 | | Crystal structure of Duvenhage virus phosphoprotein C-terminal domain | | Descriptor: | Phosphoprotein | | Authors: | Sugiyama, A, Jiang, X, Maenaka, K, Yao, M, Ose, T. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural comparison of the C-terminal domain of functionally divergent lyssavirus P proteins.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
7CMC
 
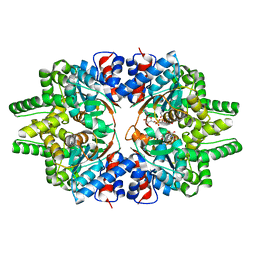 | | CRYSTAL STRUCTURE OF DEOXYHYPUSINE SYNTHASE FROM PYROCOCCUS HORIKOSHII | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable deoxyhypusine synthase | | Authors: | Yu, J, Gai, Z.Q, Okada, C, Yao, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexible NAD+Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A.
Int J Mol Sci, 21, 2020
|
|
3AJ2
 
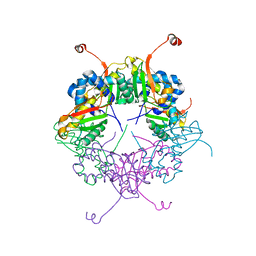 | | The structure of AxCeSD octamer (C-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2D74
 
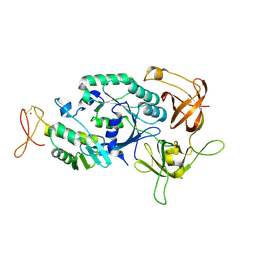 | | Crystal structure of translation initiation factor aIF2betagamma heterodimer | | Descriptor: | Translation initiation factor 2 beta subunit, Translation initiation factor 2 gamma subunit, ZINC ION | | Authors: | Sokabe, M, Yao, M, Sakai, N, Toya, S, Tanaka, I. | | Deposit date: | 2005-11-16 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3AJ1
 
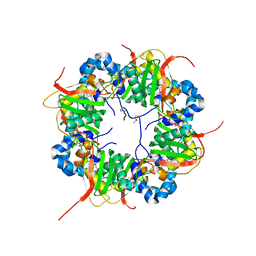 | | The structure of AxCeSD octamer (N-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2DGJ
 
 | | Crystal structure of EbhA (756-1003 domain) from Staphylococcus aureus | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tanaka, Y, Yao, M, Kuroda, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus
Structure, 16, 2008
|
|
3A8E
 
 | | The structure of AxCesD octamer complexed with cellopentaose | | Descriptor: | Cellulose synthase operon protein D, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Yao, M, Tanaka, I. | | Deposit date: | 2009-10-05 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2CVI
 
 | | Crystal structure of hypothetical protein PHS023 from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 75aa long hypothetical regulatory protein AsnC, POTASSIUM ION | | Authors: | Kitago, Y, Sakai, N, Matsumoto, D, Yao, M, Aizawa, T, Demura, M, Kawano, K, Nitta, K, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-06-03 | | Release date: | 2006-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of hypothetical protein PHS023 from Pyrococcus horikoshii
TO BE PUBLISHED
|
|
8HRH
 
 | | SN-131/1B2 anti-MUC1 antibody with a glycopeptide | | Descriptor: | 1-ACETYL-L-PROLINE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALANINE, ... | | Authors: | Wakui, H, Horidome, C, Yao, M, Ose, T, Nishimura, S.-I. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and molecular insight into antibody recognition of dynamic neoepitopes in membrane tethered MUC1 of pancreatic cancer cells and secreted exosomes.
Rsc Chem Biol, 4, 2023
|
|
8H25
 
 | | Lacticaseibacillus casei GH35 beta-galactosidase LBCZ_0230 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saburi, W, Ota, T, Kato, K, Tagami, T, Yamashita, K, Yao, M, Mori, H. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Function and Structure of Lacticaseibacillus casei GH35 beta-Galactosidase LBCZ_0230 with High Hydrolytic Activity to Lacto- N -biose I and Galacto- N -biose.
J Appl Glycosci (1999), 70, 2023
|
|
2DI3
 
 | |
1IXL
 
 | | Crystal structure of uncharacterized protein PH1136 from Pyrococcus horikoshii | | Descriptor: | hypothetical protein PH1136 | | Authors: | Tajika, Y, Sakai, N, Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2002-06-27 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of conserved protein PH1136 from Pyrococcus horikoshii.
Proteins, 55, 2004
|
|
1J08
 
 | | Crystal structure of glutaredoxin-like protein from Pyrococcus horikoshii | | Descriptor: | glutaredoxin-like protein | | Authors: | Tanaka, Y, Tanabe, E, Tsumoto, K, Kumagai, I, Yao, M, Tanaka, I. | | Deposit date: | 2002-11-11 | | Release date: | 2003-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein disulfide isomerase from hyperthermophile as an additives of refolding of an immunoglobulin-folded protein
To be Published
|
|
1J31
 
 | | Crystal Structure of Hypothetical Protein PH0642 from Pyrococcus horikoshii | | Descriptor: | ACETATE ION, Hypothetical protein PH0642 | | Authors: | Sakai, N, Tajika, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-01-16 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of hypothetical protein PH0642 from Pyrococcus horikoshii at 1.6A resolution.
Proteins, 57, 2004
|
|
1IXK
 
 | | Crystal Structure Analysis of Methyltransferase Homolog Protein from Pyrococcus Horikoshii | | Descriptor: | Methyltransferase | | Authors: | Ishikawa, I, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2002-06-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human p120 homologue protein PH1374 from Pyrococcus horikoshii
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
1J2V
 
 | | Crystal Structure of CutA1 from Pyrococcus Horikoshii | | Descriptor: | 102AA long hypothetical periplasmic divalent cation tolerance protein CUTA | | Authors: | Tanaka, Y, Sakai, N, Yasutake, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: the case of Pyrococcus horikoshii CutA.
Febs Lett., 556, 2004
|
|
1UD9
 
 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii | | Descriptor: | DNA polymerase sliding clamp A, ZINC ION | | Authors: | Tanabe, E, Yasutake, Y, Tanaka, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii
To be published
|
|
1UC2
 
 | | Hypothetical Extein Protein of PH1602 from Pyrococcus horikoshii | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, hypothetical protein PH1602 | | Authors: | Okada, C, Maegawa, Y, Yao, M, Tanaka, I. | | Deposit date: | 2003-04-08 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold
Proteins, 63, 2006
|
|
8JHH
 
 | | Glycoside hydrolase family 55 endo-beta-1,3-glucanase from Microdochium nivale | | Descriptor: | GLYCEROL, MnLam55A | | Authors: | Ota, T, Saburi, W, Yamashita, K, Tagami, T, Yu, J, Komba, S, Jewell, L.E, Hsiang, T, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2023-05-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for endo-type action of glycoside hydrolase family 55 endo-beta-1,3-glucanase on beta 1-3/1-6-glucan.
J.Biol.Chem., 299, 2023
|
|
3A24
 
 | | Crystal structure of BT1871 retaining glycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, alpha-galactosidase | | Authors: | Okuyama, M, Kitamura, M, Hondoh, H, Tanaka, I, Yao, M. | | Deposit date: | 2009-04-28 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic mechanism of retaining alpha-galactosidase belonging to glycoside hydrolase family 97.
J.Mol.Biol., 392, 2009
|
|
