7Y8Q
 
 | | Amyloid-beta assemblage on GM1-containing membranes | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yagi-Utsumi, M, Itoh, S.G, Okumura, H, Yanagisawa, K, Kato, K, Nishimura, K. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The Double-Layered Structure of Amyloid-beta Assemblage on GM1-Containing Membranes Catalytically Promotes Fibrillization.
Acs Chem Neurosci, 14, 2023
|
|
1KVB
 
 | | E. COLI RIBONUCLEASE HI D134H MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVA
 
 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVC
 
 | | E. COLI RIBONUCLEASE HI D134N MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
5GKI
 
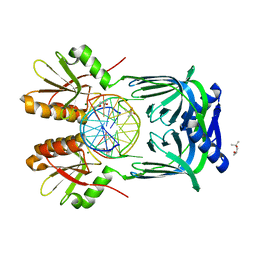 | | Structure of EndoMS-dsDNA3 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*CP*TP*AP*GP*GP*TP*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*GP*GP*GP*CP*CP*TP*AP*GP*GP*C)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKG
 
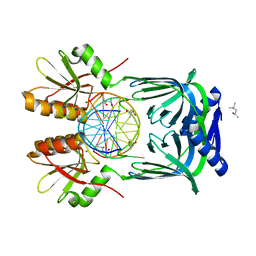 | | Structure of EndoMS-dsDNA1'' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*GP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKF
 
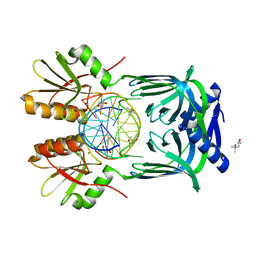 | | Structure of EndoMS-dsDNA1' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*TP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*TP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKE
 
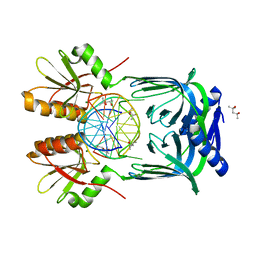 | | Structure of EndoMS-dsDNA1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*TP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKH
 
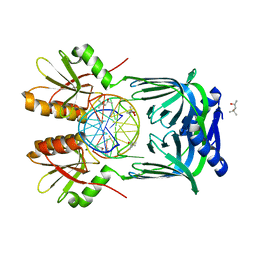 | | Structure of EndoMS-dsDNA2 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*TP*TP*GP*GP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*CP*CP*AP*GP*GP*TP*GP*CP*CP*GP*T)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKJ
 
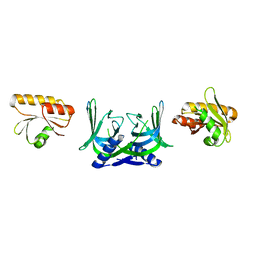 | | Structure of EndoMS in apo form | | Descriptor: | Endonuclease EndoMS | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
1RIL
 
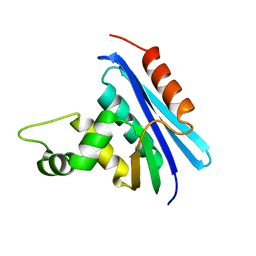 | | CRYSTAL STRUCTURE OF RIBONUCLEASE H FROM THERMUS THERMOPHILUS HB8 REFINED AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Okumura, M, Katayanagi, K, Kimura, S, Kanaya, S, Nakamura, H, Morikawa, K. | | Deposit date: | 1993-01-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ribonuclease H from Thermus thermophilus HB8 refined at 2.8 A resolution.
J.Mol.Biol., 230, 1993
|
|
1ENK
 
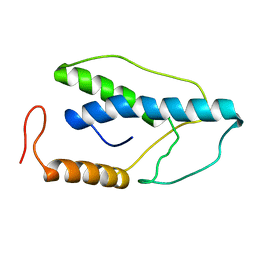 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENI
 
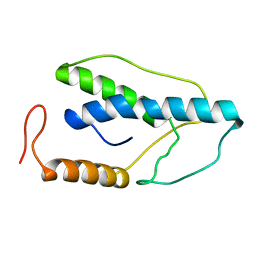 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
3B0L
 
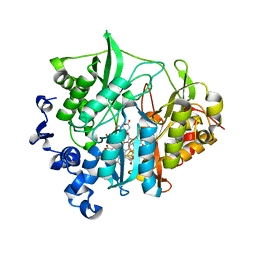 | | M175G mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B0M
 
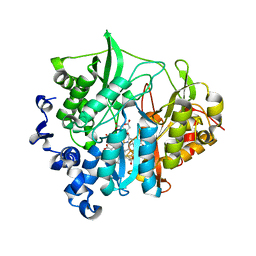 | | M175K mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B0N
 
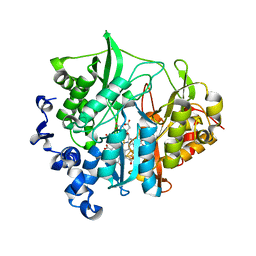 | | Q448K mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B0G
 
 | | Assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B0H
 
 | | Assimilatory nitrite reductase (Nii4) from tobbaco root | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B0J
 
 | | M175E mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
1ENJ
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
2ZSY
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [750 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZSR
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [450 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZT0
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 100 K: Laser on [750 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZT1
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 100 K: Laser on [810 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
