1MPT
 
 | | CRYSTAL STRUCTURE OF A NEW ALKALINE SERINE PROTEASE (M-PROTEASE) FROM BACILLUS SP. KSM-K16 | | 分子名称: | CALCIUM ION, M-PROTEASE | | 著者 | Yamane, T, Kani, T, Hatanaka, T, Suzuki, A, Ashida, T, Kobayashi, T, Ito, S, Yamashita, O. | | 登録日 | 1994-04-13 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of a new alkaline serine protease (M-protease) from Bacillus sp. KSM-K16.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1JWQ
 
 | | Structure of the catalytic domain of CwlV, N-acetylmuramoyl-L-alanine amidase from Bacillus(Paenibacillus) polymyxa var.colistinus | | 分子名称: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE CwlV, ZINC ION | | 著者 | Yamane, T, Koyama, Y, Nojiri, Y, Hikage, T, Akita, M, Suzuki, A, Shirai, T, Ise, F, Shida, T, Sekiguchi, J. | | 登録日 | 2001-09-05 | | 公開日 | 2003-11-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Structure of the catalytic domain of N-acetylmuramoyl-L-alanine amidase, a cell wall hydrolase from Bacillus polymyxa var.colistinus and its resemblance to the structure of carboxypeptidases
To be Published
|
|
2Z33
 
 | | Solution structure of the DNA complex of PhoB DNA-binding/transactivation Domain | | 分子名称: | 5'-D(*AP*CP*AP*GP*AP*TP*TP*TP*AP*TP*GP*AP*CP*AP*GP*T)-3', 5'-D(*AP*CP*TP*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*TP*GP*T)-3', Phosphate regulon transcriptional regulatory protein phoB | | 著者 | Yamane, T, Okamura, H, Ikeguchi, M, Nishimura, Y, Kidera, A. | | 登録日 | 2007-05-31 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment.
Proteins, 71, 2008
|
|
3DXW
 
 | | The crystal structure of alpha-amino-epsilon-caprolactam racemase from Achromobacter obae complexed with epsilon caprolactam | | 分子名称: | Alpha-amino-epsilon-caprolactam racemase, PYRIDOXAL-5'-PHOSPHATE, azepan-2-one | | 著者 | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | 登録日 | 2008-07-25 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | The novel structure of a pyridoxal 5'-phosphate-dependent fold-type I racemase, alpha-amino-epsilon-caprolactam racemase from Achromobacter obae
Biochemistry, 48, 2009
|
|
3DXV
 
 | | The crystal structure of alpha-amino-epsilon-caprolactam racemase from Achromobacter obae | | 分子名称: | Alpha-amino-epsilon-caprolactam racemase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Okazaki, S, Suzuki, A, Komeda, H, Asano, Y, Yamane, T. | | 登録日 | 2008-07-25 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | The novel structure of a pyridoxal 5'-phosphate-dependent fold-type I racemase, alpha-amino-epsilon-caprolactam racemase from Achromobacter obae
Biochemistry, 48, 2009
|
|
1QM8
 
 | | Structure of Bacteriorhodopsin at 100 K | | 分子名称: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | 著者 | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Kamiya, N, Kouyama, T. | | 登録日 | 1999-09-22 | | 公開日 | 2000-08-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A Novel Three-Dimensional Crystal of Bacteriorhodopsin Obtained by Successive Fusion of the Vesicular Assemblies.
J.Mol.Biol., 283, 1998
|
|
1CYC
 
 | | THE CRYSTAL STRUCTURE OF BONITO (KATSUO) FERROCYTOCHROME C AT 2.3 ANGSTROMS RESOLUTION. II. STRUCTURE AND FUNCTION | | 分子名称: | FERROCYTOCHROME C, HEME C | | 著者 | Tanaka, N, Yamane, T, Tsukihara, T, Ashida, T, Kakudo, M. | | 登録日 | 1976-08-01 | | 公開日 | 1976-10-06 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
J.Biochem.(Tokyo), 77, 1975
|
|
2RJ2
 
 | | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution | | 分子名称: | CHLORIDE ION, F-box only protein 2, NICKEL (II) ION | | 著者 | Vaijayanthimala, S, Velmurugan, D, Mizushima, T, Yamane, T, Yoshida, Y, Tanaka, K. | | 登録日 | 2007-10-14 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution
To be Published
|
|
2RPQ
 
 | | Solution Structure of a SUMO-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3 | | 分子名称: | Activating transcription factor 7-interacting protein 1, Small ubiquitin-related modifier 2 | | 著者 | Sekiyama, N, Ikegami, T, Yamane, T, Ikeguchi, M, Uchimura, Y, Baba, D, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | 登録日 | 2008-07-07 | | 公開日 | 2008-10-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3
J.Biol.Chem., 283, 2008
|
|
3HJE
 
 | | Crystal structure of sulfolobus tokodaii hypothetical maltooligosyl trehalose synthase | | 分子名称: | 704aa long hypothetical glycosyltransferase, GLYCEROL | | 著者 | Cielo, C.B.C, Okazaki, S, Suzuki, A, Mizushima, T, Masui, R, Kuramitsu, S, Yamane, T. | | 登録日 | 2009-05-21 | | 公開日 | 2010-04-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of ST0929, a putative glycosyl transferase from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2RVQ
 
 | | Solution structure of the isolated histone H2A-H2B heterodimer | | 分子名称: | Histone H2A type 1-B/E, Histone H2B type 1-J | | 著者 | Moriwaki, Y, Yamane, T, Ohtomo, H, Ikeguchi, M, Kurita, J, Sato, M, Nagadoi, A, Shimojo, H, Nishimura, Y. | | 登録日 | 2016-03-28 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the isolated histone H2A-H2B heterodimer
Sci Rep, 6, 2016
|
|
2GGS
 
 | | crystal structure of hypothetical dTDP-4-dehydrorhamnose reductase from sulfolobus tokodaii | | 分子名称: | 273aa long hypothetical dTDP-4-dehydrorhamnose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | 登録日 | 2006-03-24 | | 公開日 | 2007-03-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | crystal structure of hypothetical dTDP-4-dehydrorhamnose reductase from sulfolobus tokodaii
To be published
|
|
2GGQ
 
 | | complex of hypothetical glucose-1-phosphate thymidylyltransferase from sulfolobus tokodaii | | 分子名称: | 401aa long hypothetical glucose-1-phosphate thymidylyltransferase, IODIDE ION, THYMIDINE-5'-TRIPHOSPHATE | | 著者 | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | 登録日 | 2006-03-24 | | 公開日 | 2007-03-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | complex of hypothetical glucose-1-phosphate thymidylyltransferase from sulfolobus tokodaii
To be published
|
|
2GGO
 
 | | Crystal Structure of glucose-1-phosphate thymidylyltransferase from Sulfolobus tokodaii | | 分子名称: | 401aa long hypothetical glucose-1-phosphate thymidylyltransferase | | 著者 | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | 登録日 | 2006-03-24 | | 公開日 | 2007-03-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of glucose-1-phosphate thymidylyltransferase from
Sulfolobus tokodaii
To be published
|
|
1C1F
 
 | | LIGAND-FREE CONGERIN I | | 分子名称: | PROTEIN (CONGERIN I) | | 著者 | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | 登録日 | 1999-03-03 | | 公開日 | 1999-10-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1C1L
 
 | | LACTOSE-LIGANDED CONGERIN I | | 分子名称: | PROTEIN (CONGERIN I), beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Shirai, T, Mitsuyama, C, Niwa, Y, Matsui, Y, Hotta, H, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | 登録日 | 1999-03-03 | | 公開日 | 1999-10-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | High-resolution structure of the conger eel galectin, congerin I, in lactose-liganded and ligand-free forms: emergence of a new structure class by accelerated evolution.
Structure Fold.Des., 7, 1999
|
|
1CH4
 
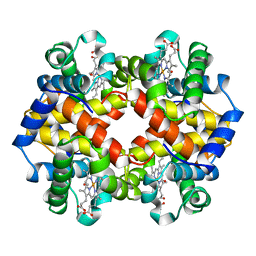 | | MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA (F133V) | | 分子名称: | CARBON MONOXIDE, MODULE-SUBSTITUTED CHIMERA HEMOGLOBIN BETA-ALPHA, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Shirai, T, Fujikake, M, Yamane, T, Inaba, K, Ishimori, K, Morishima, I. | | 登録日 | 1998-06-11 | | 公開日 | 1999-04-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a protein with an artificial exon-shuffling, module M4-substituted chimera hemoglobin beta alpha, at 2.5 A resolution.
J.Mol.Biol., 287, 1999
|
|
2ZE4
 
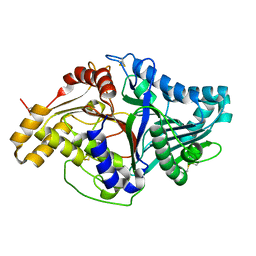 | | Crystal structure of phospholipase D from streptomyces antibioticus | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Phospholipase D | | 著者 | Suzuki, A, Kakuno, K, Saito, R, Iwasaki, Y, Yamane, T, Yamane, T. | | 登録日 | 2007-12-05 | | 公開日 | 2007-12-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of phospholipase D from streptomyces antibioticus
To be Published
|
|
2ZE9
 
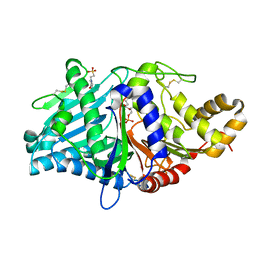 | | Crystal structure of H168A mutant of phospholipase D from Streptomyces antibioticus, as a complex with phosphatidylcholine | | 分子名称: | (2R)-3-(phosphonooxy)propane-1,2-diyl diheptanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Phospholipase D | | 著者 | Suzuki, A, Toda, H, Iwasaki, Y, Yamane, T, Yamane, T. | | 登録日 | 2007-12-06 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of phospholipase D from streptomyces antibioticus
To be Published
|
|
1J3F
 
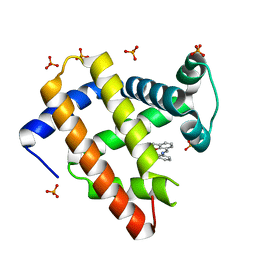 | | Crystal Structure of an Artificial Metalloprotein:Cr(III)(3,3'-Me2-salophen)/apo-A71G Myoglobin | | 分子名称: | 'N,N'-BIS-(2-HYDROXY-3-METHYL-BENZYLIDENE)-BENZENE-1,2-DIAMINE', CHROMIUM ION, Myoglobin, ... | | 著者 | Koshiyama, T, Kono, M, Ohashi, M, Ueno, T, Suzuki, A, Yamane, T, Watanabe, Y. | | 登録日 | 2003-01-24 | | 公開日 | 2004-05-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Coordinated Design of Cofactor and Active Site Structures in Development of New Protein Catalysts
J.Am.Chem.Soc., 127, 2005
|
|
3WSO
 
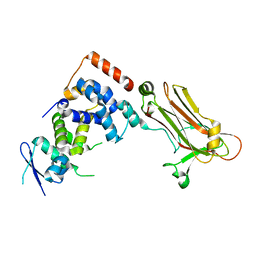 | | Crystal structure of the Skp1-FBG3 complex | | 分子名称: | F-box only protein 44, S-phase kinase-associated protein 1 | | 著者 | Kumanomidou, T, Nishio, K, Takagi, K, Nakagawa, T, Suzuki, A, Yamane, T, Tokunaga, F, Iwai, K, Murakami, A, Yoshida, Y, Tanaka, K, Mizushima, T. | | 登録日 | 2014-03-18 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Structural Differences between a Glycoprotein Specific F-Box Protein Fbs1 and Its Homologous Protein FBG3
Plos One, 10, 2015
|
|
1EE6
 
 | | CRYSTAL STRUCTURE OF PECTATE LYASE FROM BACILLUS SP. STRAIN KSM-P15. | | 分子名称: | CALCIUM ION, PECTATE LYASE | | 著者 | Akita, M, Suzuki, A, Kobayashi, T, Ito, S, Yamane, T. | | 登録日 | 2000-01-31 | | 公開日 | 2001-01-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The first structure of pectate lyase belonging to polysaccharide lyase family 3.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G0C
 
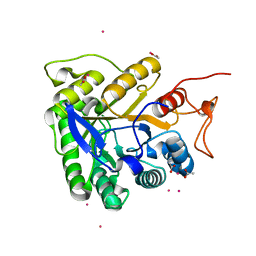 | | ALKALINE CELLULASE K CATALYTIC DOMAIN-CELLOBIOSE COMPLEX | | 分子名称: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE, ... | | 著者 | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | 登録日 | 2000-10-05 | | 公開日 | 2001-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
1G01
 
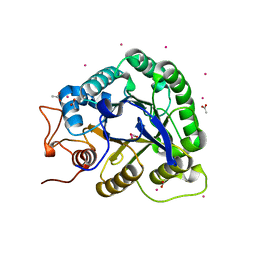 | | ALKALINE CELLULASE K CATALYTIC DOMAIN | | 分子名称: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE | | 著者 | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | 登録日 | 2000-10-05 | | 公開日 | 2001-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
3X3H
 
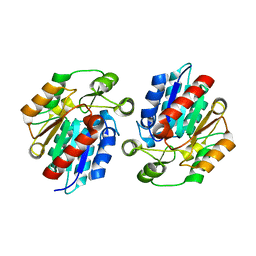 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) 3KP (K176P, K199P, K224P) triple mutant | | 分子名称: | (S)-hydroxynitrile lyase | | 著者 | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H, Okazaki, S. | | 登録日 | 2015-01-21 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
TO BE PUBLISHED
|
|
