5EJB
 
 | | Crystal structure of prefusion Hendra virus F protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Wong, J.W, Jardetzky, T.S, Paterson, R.G, Lamb, R.A. | | Deposit date: | 2015-11-01 | | Release date: | 2016-01-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and stabilization of the Hendra virus F glycoprotein in its prefusion form.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3OMY
 
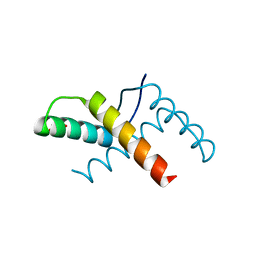 | | Crystal structure of the pED208 TraM N-terminal domain | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein traM | | Authors: | Wong, J.J.W, Lu, J, Edwards, R.A, Frost, L.S, Mark Glover, J.N. | | Deposit date: | 2010-08-27 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM.
Nucleic Acids Res., 39, 2011
|
|
3ON0
 
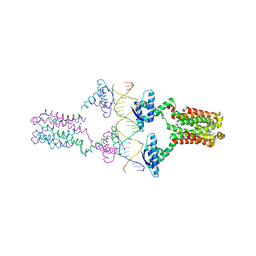 | | Crystal structure of the pED208 TraM-sbmA complex | | Descriptor: | Protein traM, sbmA | | Authors: | Wong, J.J.W, Lu, J, Edwards, R.A, Frost, L.S, Mark Glover, J.N. | | Deposit date: | 2010-08-27 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.874 Å) | | Cite: | Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM.
Nucleic Acids Res., 39, 2011
|
|
4E3R
 
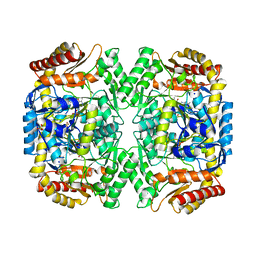 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4E3Q
 
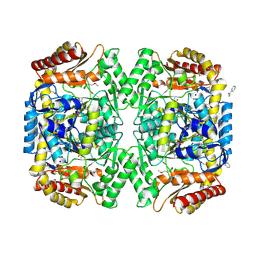 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
