5HQH
 
 | | 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes. | | Descriptor: | CHLORIDE ION, Lmo2119 protein, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes.
To Be Published
|
|
2L3M
 
 | | Solution structure of the putative copper-ion-binding protein from Bacillus anthracis str. Ames | | Descriptor: | Copper-ion-binding protein | | Authors: | Zhang, Y, Winsor, J, Dubrovska, I, Anderson, W, Radhakrishnan, I, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-16 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | To be published
To be Published
|
|
3LAY
 
 | | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | Zinc resistance-associated protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-07 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
5VGM
 
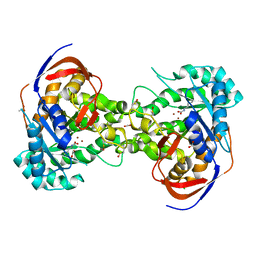 | | Crystal structure of dihydroorotase pyrC from Vibrio cholerae in complex with zinc at 1.95 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Lipowska, J, Shabalin, I.G, Miks, C.D, Winsor, J, Cooper, D.R, Shuvalova, L, Kwon, K, Lewinski, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-11 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
3V05
 
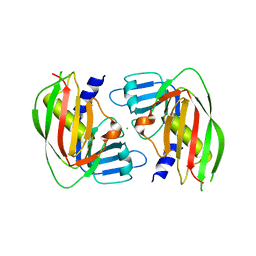 | | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Superantigen-like Protein | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Filippova, E.V, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3UWQ
 
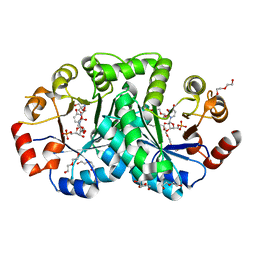 | | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Shuvalova, L, Kuhn, M, Filippova, E.V, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP)
To be Published
|
|
3KY7
 
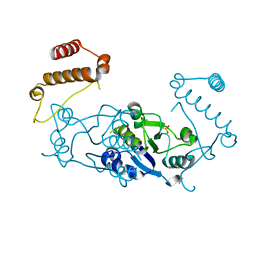 | | 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252 | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, See, R, Zoraghi, R, Reiner, N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-04 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of a putative tRNA (guanine-7-)-methyltransferase (trmD) from Staphylococcus aureus subsp. aureus MRSA252
To be Published
|
|
3DZC
 
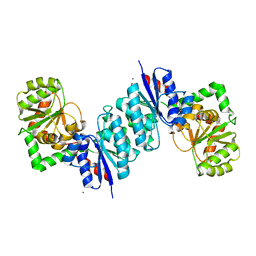 | | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Kwon, K, Hasseman, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae.
TO BE PUBLISHED
|
|
3URY
 
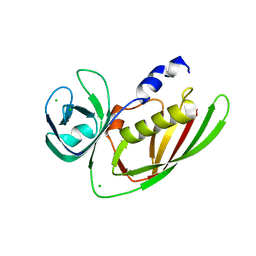 | | Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus subsp. aureus NCTC 8325 | | Descriptor: | CHLORIDE ION, Exotoxin | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus subsp. aureus NCTC 8325
To be Published
|
|
3UN6
 
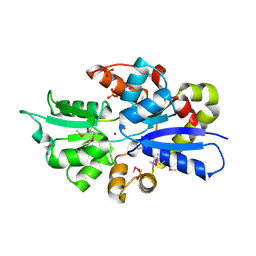 | | 2.0 Angstrom Crystal Structure of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Zinc bound | | Descriptor: | ABC transporter substrate-binding protein, PHOSPHATE ION, ZINC ION | | Authors: | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-15 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2.0 Angstrom Crystal Structure of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Zinc bound.
TO BE PUBLISHED
|
|
3KZW
 
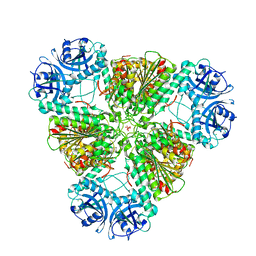 | | Crystal structure of cytosol aminopeptidase from Staphylococcus aureus COL | | Descriptor: | CHLORIDE ION, Cytosol aminopeptidase, PHOSPHATE ION, ... | | Authors: | Hattne, J, Dubrovska, I, Halavaty, A, Minasov, G, Scott, P, Shuvalova, L, Winsor, J, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: |
|
|
2LRJ
 
 | |
2M1Z
 
 | |
6BWT
 
 | | 2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase from Francisella tularensis. | | Descriptor: | CHLORIDE ION, SULFATE ION, Thioredoxin reductase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure Thioredoxin Reductase from Francisella tularensis.
To Be Published
|
|
6C43
 
 | | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium. | | Descriptor: | Gamma-aminobutyraldehyde dehydrogenase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Tekleab, H, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-11 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium.
To Be Published
|
|
5UTU
 
 | | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD
To Be Published
|
|
5US8
 
 | | 2.15 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis
To Be Published
|
|
5UJS
 
 | | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.
To Be Published
|
|
5I0C
 
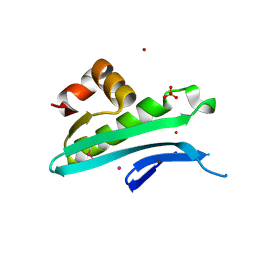 | | Crystal structure of predicted acyltransferase YjdJ with acyl-CoA N-acyltransferase domain from Escherichia coli str. K-12 | | Descriptor: | CADMIUM ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-03 | | Release date: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of predicted acyltransferase YjdJ with acyl-CoA N-acyltransferase domain from Escherichia coli str. K-12
To Be Published
|
|
3VCZ
 
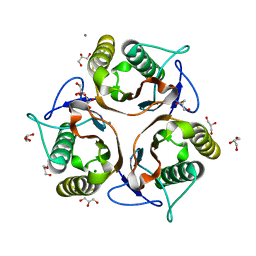 | | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6 | | Descriptor: | CALCIUM ION, Endoribonuclease L-PSP, GLYCEROL, ... | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E.V, Dubrovska, I, Winsor, J, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of a putative translation initiation inhibitor from Vibrio vulnificus CMCP6
To be Published
|
|
5IKM
 
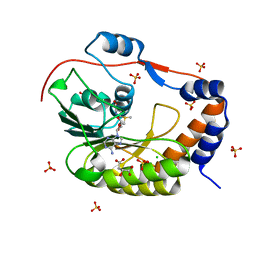 | | 1.9 Angstrom Crystal Structure of NS5 Methyl Transferase from Dengue Virus 1 in Complex with S-Adenosylmethionine and Beta-D-Fructopyranose. | | Descriptor: | CHLORIDE ION, D-MALATE, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Flores, K, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-03 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of NS5 Methyl Transferase from Dengue Virus 1 in Complex with S-Adenosylmethionine and Beta-D-Fructopyranose.
To Be Published
|
|
4QTO
 
 | | 1.65 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-modified Cys289 and PEG molecule in active site | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3V4H
 
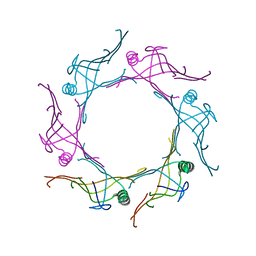 | | Crystal structure of a type VI secretion system effector from Yersinia pestis | | Descriptor: | hypothetical protein | | Authors: | Filippova, E.V, Halavaty, A, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a type VI secretion system effector from Yersinia pestis
To be Published
|
|
5TV7
 
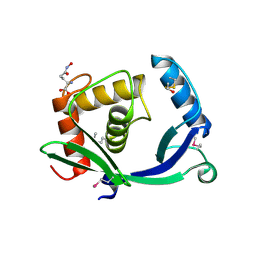 | | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate. | | Descriptor: | GLUTAMINE HYDROXAMATE, Putative peptidoglycan-binding/hydrolysing protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Winsor, J, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate.
To Be Published
|
|
3V4G
 
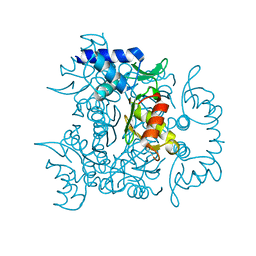 | | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6 | | Descriptor: | Arginine repressor | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E, Shuvalova, L, Winsor, J, Dubrovska, I, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6
To be Published
|
|
