4RVE
 
 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*TP*AP*TP*CP*CP*C)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilso, K.S. | | Deposit date: | 1993-02-18 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
3MHY
 
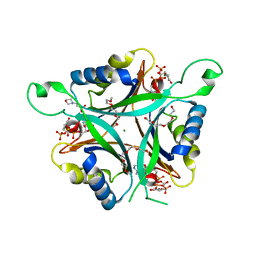 | | A New PII Protein Structure | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ... | | Authors: | Winkler, F.K, Truan, D, Li, X.D. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A new P(II) protein structure identifies the 2-oxoglutarate binding site.
J.Mol.Biol., 400, 2010
|
|
2RVE
 
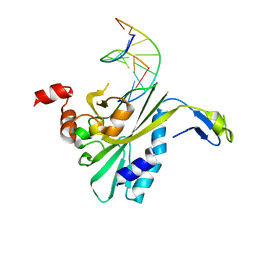 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*CP*TP*CP*G)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilson, K.S. | | Deposit date: | 1991-03-19 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
1RVE
 
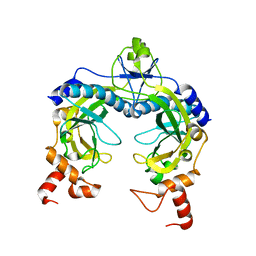 | |
2TCL
 
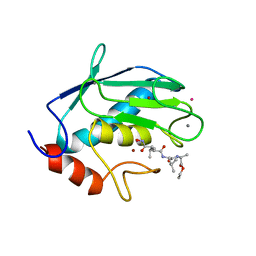 | | STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH AN INHIBITOR | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, SAMARIUM (III) ION, ... | | Authors: | Winkler, F.K, Borkakoti, N, D'Arcy, A. | | Deposit date: | 1994-09-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the catalytic domain of human fibroblast collagenase complexed with an inhibitor.
Nat.Struct.Biol., 1, 1994
|
|
1JRH
 
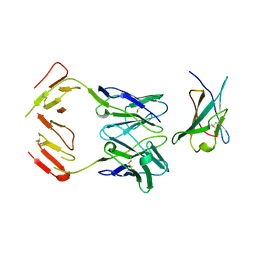 | | COMPLEX (ANTIBODY/ANTIGEN) | | Descriptor: | ANTIBODY A6, INTERFERON-GAMMA RECEPTOR ALPHA CHAIN | | Authors: | Winkler, F.K, Sogabe, S. | | Deposit date: | 1997-09-23 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neutralizing epitopes on the extracellular interferon gamma receptor (IFNgammaR) alpha-chain characterized by homolog scanning mutagenesis and X-ray crystal structure of the A6 fab-IFNgammaR1-108 complex.
J.Mol.Biol., 273, 1997
|
|
3B9W
 
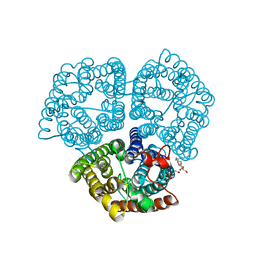 | |
3O5T
 
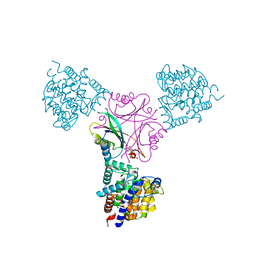 | | Structure of DraG-GlnZ complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dinitrogenase reductase activacting glicohydrolase, MAGNESIUM ION, ... | | Authors: | Rajendran, C, Li, X.-D, Winkler, F.K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of the GlnZ-DraG complex reveals a different form of PII-target interaction
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1RVC
 
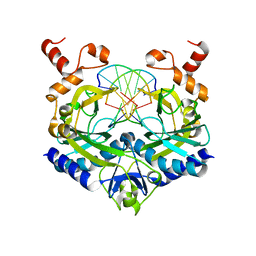 | |
1RVB
 
 | |
1RVA
 
 | |
2AKF
 
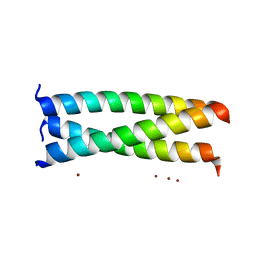 | | Crystal structure of the coiled-coil domain of coronin 1 | | Descriptor: | Coronin-1A, ZINC ION | | Authors: | Kammerer, R.A, Kostrewa, D, Progias, P, Honnappa, S, Avila, D, Lustig, A, Winkler, F.K, Pieters, J, Steinmetz, M.O. | | Deposit date: | 2005-08-03 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A conserved trimerization motif controls the topology of short coiled coils
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3G9D
 
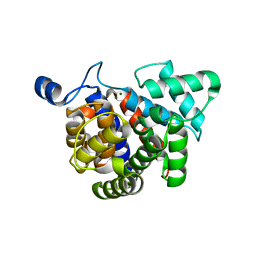 | | Crystal structure glycohydrolase | | Descriptor: | Dinitrogenase reductase activating glucohydrolase, MAGNESIUM ION | | Authors: | Li, X.-D, Winkler, F.K. | | Deposit date: | 2009-02-13 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Dinitrogenase Reductase-activating Glycohydrolase (DRAG) Reveals Conservation in the ADP-Ribosylhydrolase Fold and Specific Features in the ADP-Ribose-binding Pocket
J.Mol.Biol., 390, 2009
|
|
2GNN
 
 | | Crystal Structure of the Orf Virus NZ2 Variant of VEGF-E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, ... | | Authors: | Prota, A.E, Pieren, M, Wagner, A, Kostrewa, D, Winkler, F.K, Ballmer-Hofer, K. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Orf virus NZ2 variant of vascular endothelial growth factor-E. Implications for receptor specificity.
J.Biol.Chem., 281, 2006
|
|
2HL5
 
 | |
2HL3
 
 | |
1F97
 
 | | SOLUBLE PART OF THE JUNCTION ADHESION MOLECULE FROM MOUSE | | Descriptor: | JUNCTION ADHESION MOLECULE, MAGNESIUM ION | | Authors: | Kostrewa, D, Brockhaus, M, D'Arcy, A, Dale, G, Bazzoni, G, Dejana, E, Winkler, F, Hennig, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif.
EMBO J., 20, 2001
|
|
2HKN
 
 | |
2HKQ
 
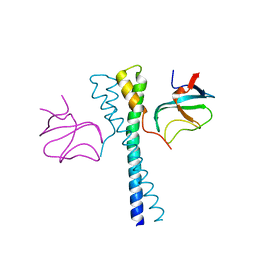 | |
1Z6B
 
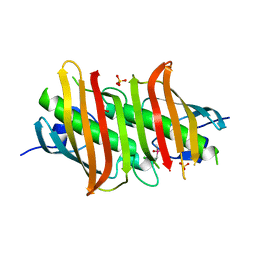 | | Crystal structure of Plasmodium falciparum FabZ at 2.1 A | | Descriptor: | CACODYLATE ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kostrewa, D, Winkler, F.K, Folkers, G, Scapozza, L, Perozzo, R. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum
PROTEIN SCI., 14, 2005
|
|
1XQP
 
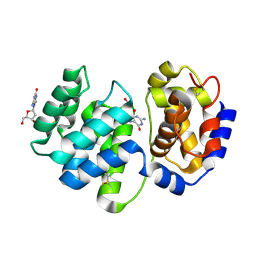 | | Crystal structure of 8-oxoguanosine complexed Pa-AGOG, 8-oxoguanine DNA glycosylase from Pyrobaculum aerophilum | | Descriptor: | 2'-DEOXY-8-OXOGUANOSINE, 8-oxoguanine DNA glycosylase | | Authors: | Lingaraju, G.M, Sartori, A.A, Kostrewa, D, Prota, A.E, Jiricny, J, Winkler, F.K. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure
Structure, 13, 2005
|
|
1XQO
 
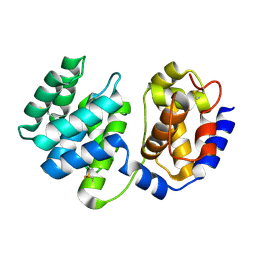 | | Crystal structure of native Pa-AGOG, 8-oxoguanine DNA glycosylase from Pyrobaculum aerophilum | | Descriptor: | 8-oxoguanine DNA glycosylase | | Authors: | Lingaraju, G.M, Sartori, A.A, Kostrewa, D, Prota, A.E, Jiricny, J, Winkler, F.K. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure
Structure, 13, 2005
|
|
3C1J
 
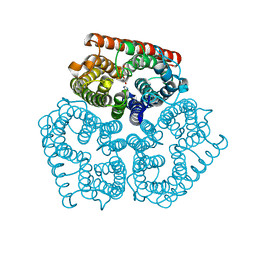 | | Substrate binding, deprotonation and selectivity at the periplasmic entrance of the E. coli ammonia channel AmtB | | Descriptor: | ACETATE ION, Ammonia channel, GLYCEROL, ... | | Authors: | Lupo, D, Winkler, F.K. | | Deposit date: | 2008-01-23 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C1G
 
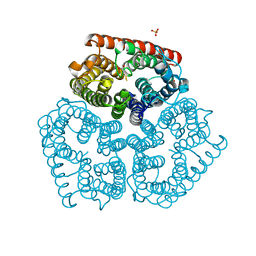 | | Substrate binding, deprotonation and selectivity at the periplasmic entrance of the E. coli ammonia channel AmtB | | Descriptor: | ACETATE ION, Ammonia channel, SULFATE ION | | Authors: | Javelle, A, Kostrewa, D, Lupo, D, Winkler, F.K. | | Deposit date: | 2008-01-23 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C1I
 
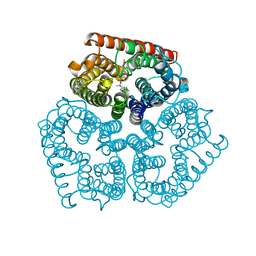 | | Substrate binding, deprotonation and selectivity at the periplasmic entrance of the E. coli ammonia channel AmtB | | Descriptor: | ACETATE ION, Ammonia channel, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Lupo, D, Winkler, F.K. | | Deposit date: | 2008-01-23 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
