2AB0
 
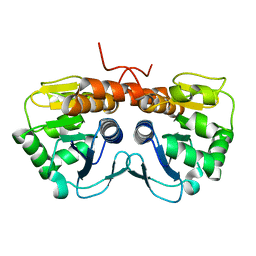 | | Crystal Structure of E. coli protein YajL (ThiJ) | | Descriptor: | YajL | | Authors: | Wilson, M.A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-07-14 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Atomic Resolution Crystal Structure of the YajL (ThiJ) Protein from Escherichia coli: A Close Prokaryotic Homologue of the Parkinsonism-associated Protein DJ-1.
J.Mol.Biol., 353, 2005
|
|
6NIA
 
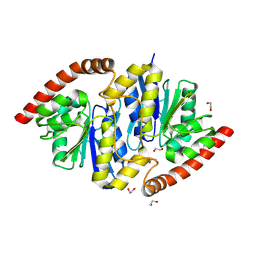 | |
6NI9
 
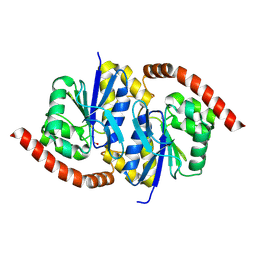 | |
6NI5
 
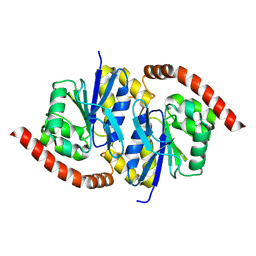 | |
6NI6
 
 | |
6NI4
 
 | |
6NI8
 
 | |
6NI7
 
 | |
8TSY
 
 | |
8TT0
 
 | |
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSZ
 
 | |
8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TT5
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=8.3 | | Descriptor: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2023-08-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
8TSU
 
 | |
8TSX
 
 | |
8TT1
 
 | |
8VQ1
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, thioimidate intermediate | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA, N-(4-nitrophenyl)methanimine | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
8VPW
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, free enzyme | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
2V14
 
 | | Kinesin 16B Phox-homology domain (KIF16B) | | Descriptor: | KINESIN-LIKE MOTOR PROTEIN C20ORF23 | | Authors: | Wilson, M.I, Williams, R.L, Cho, W, Hong, W, Blatner, N.R. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Novel Endosome Anchoring Activity of Kif16B Kinesin.
Embo J., 26, 2007
|
|
3GL2
 
 | | Crystal structure of dicamba monooxygenase bound to dicamba | | Descriptor: | 3,6-dichloro-2-methoxybenzoic acid, DdmC, FE (III) ION, ... | | Authors: | Wilson, M.A, Dumitru, R, Jiang, W.Z, Weeks, D.P. | | Deposit date: | 2009-03-11 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation.
J.Mol.Biol., 392, 2009
|
|
3GL0
 
 | | Crystal structure of dicamba monooxygenase bound to 3,6 dichlorosalicylic acid (DCSA) | | Descriptor: | 1,2-ETHANEDIOL, 3,6-dichloro-2-hydroxybenzoic acid, DdmC, ... | | Authors: | Wilson, M.A, Dumitru, R, Jiang, W.Z, Weeks, D.P. | | Deposit date: | 2009-03-11 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation.
J.Mol.Biol., 392, 2009
|
|
3GKE
 
 | | Crystal Structure of Dicamba Monooxygenase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DdmC, ... | | Authors: | Wilson, M.A, Dumitru, R, Jiang, W.Z, Weeks, D.P. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation.
J.Mol.Biol., 392, 2009
|
|
5KGF
 
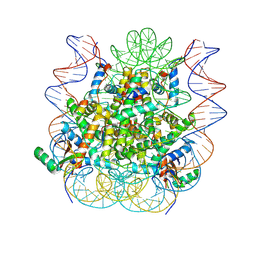 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-27 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
4QYX
 
 | | Crystal structure of YDR533Cp | | Descriptor: | Probable chaperone protein HSP31 | | Authors: | Wilson, M.A, Amour, S.T, Collins, J.L, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
