3Q09
 
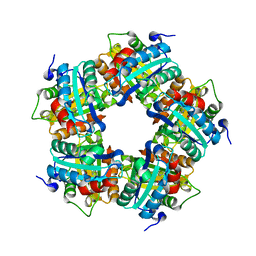 | |
3POT
 
 | |
3Q08
 
 | |
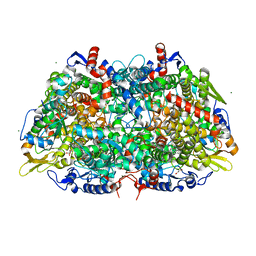
3PXW
 
 | | Crystal Structure of Ferrous NO Adduct of MauG in Complex with Pre-Methylamine Dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, ... | | Authors: | Yukl, E.T, Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structures of CO and NO Adducts of MauG in Complex with Pre-Methylamine Dehydrogenase: Implications for the Mechanism of Dioxygen Activation.
Biochemistry, 50, 2011
|
|
3PXT
 
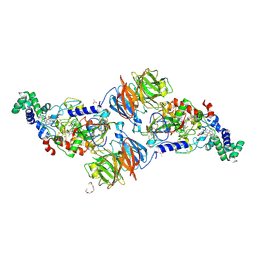 | | Crystal Structure of Ferrous CO Adduct of MauG in Complex with Pre-Methylamine Dehydrogenase | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, CALCIUM ION, ... | | Authors: | Yukl, E.T, Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of CO and NO Adducts of MauG in Complex with Pre-Methylamine Dehydrogenase: Implications for the Mechanism of Dioxygen Activation.
Biochemistry, 50, 2011
|
|
3ROW
 
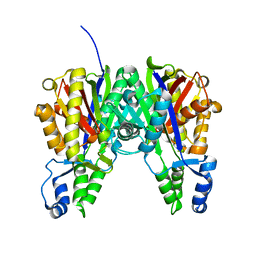 | | Crystal Structure of Xanthomonas campestri OleA | | Descriptor: | 3-oxoacyl-[ACP] synthase III | | Authors: | Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8487 Å) | | Cite: | Crystal Structures of Xanthomonas campestris OleA Reveal Features That Promote Head-to-Head Condensation of Two Long-Chain Fatty Acids.
Biochemistry, 51, 2012
|
|
3RN1
 
 | |
3RLM
 
 | |
3RMZ
 
 | |
3RN0
 
 | |
3S20
 
 | | Crystal structure of cerulenin bound Xanthomonas campestri OleA (soak) | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | Authors: | Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2011-05-16 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8796 Å) | | Cite: | Crystal Structures of Xanthomonas campestris OleA Reveal Features That Promote Head-to-Head Condensation of Two Long-Chain Fatty Acids.
Biochemistry, 51, 2012
|
|
3S1Z
 
 | |
3S21
 
 | | Crystal structure of cerulenin bound Xanthomonas campestri OleA (co-crystal) | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | Authors: | Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2011-05-16 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7001 Å) | | Cite: | Crystal Structures of Xanthomonas campestris OleA Reveal Features That Promote Head-to-Head Condensation of Two Long-Chain Fatty Acids.
Biochemistry, 51, 2012
|
|
3S23
 
 | | Crystal structure of cerulenin bound Xanthomonas campestri oleA (co-crystal) Xe Derivative | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | Authors: | Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2011-05-16 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9484 Å) | | Cite: | Crystal Structures of Xanthomonas campestris OleA Reveal Features That Promote Head-to-Head Condensation of Two Long-Chain Fatty Acids.
Biochemistry, 51, 2012
|
|
3SJL
 
 | |
3SLE
 
 | |
4Z60
 
 | |
4Z5Y
 
 | |
4Z7R
 
 | |
