2VRP
 
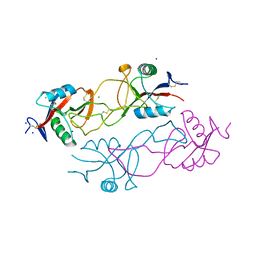 | | Structure of rhodocytin | | Descriptor: | AGGRETIN ALPHA CHAIN, AGGRETIN BETA CHAIN, CHLORIDE ION, ... | | Authors: | Watson, A.A, O'Callaghan, C.A. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of Rhodocytin, a Ligand for the Platelet-Activating Receptor Clec-2.
Protein Sci., 17, 2008
|
|
2YHF
 
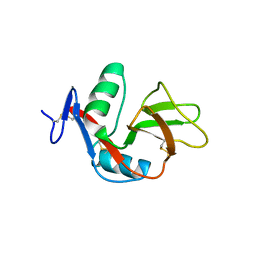 | | 1.9 Angstrom Crystal Structure of CLEC5A | | Descriptor: | C-TYPE LECTIN DOMAIN FAMILY 5 MEMBER A | | Authors: | Watson, A.A, Lebedev, A.A, Murshudov, G.M, Vagin, A.A, Hall, B.A, O'Callaghan, C.A. | | Deposit date: | 2011-04-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Flexibility of the Macrophage Dengue Virus Receptor Clec5A: Implications for Ligand Binding and Signaling.
J.Biol.Chem., 286, 2011
|
|
2C6U
 
 | |
1BA6
 
 | |
4BBQ
 
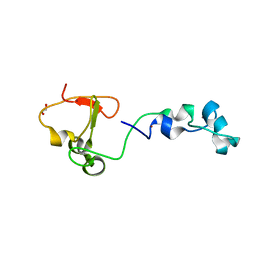 | | Crystal structure of the CXXC and PHD domain of Human Lysine-specific Demethylase 2A (KDM2A)(FBXL11) | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 2A, ZINC ION | | Authors: | Allerston, C.K, Watson, A.A, Edlich, C, Li, B, Chen, Y, Ball, L, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Laue, E.D, Gileadi, O. | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of the Cxxc and Phd Domain of Human Lysine-Specific Demethylase 2A (Kdm2A)(Fbxl11)
To be Published
|
|
4PBY
 
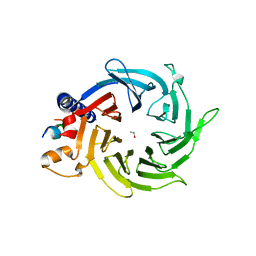 | | Structure of the human RbAp48-MTA1(656-686) complex | | Descriptor: | Histone-binding protein RBBP4, ISOPROPYL ALCOHOL, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Lejon, S, Alqarni, S.S.M, Silva, A.P.G, Watson, A.A, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
4CA9
 
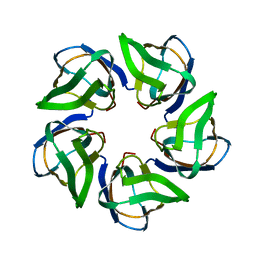 | | Structure of the Nucleoplasmin-like N-terminal domain of Drosophila FKBP39 | | Descriptor: | 39 KDA FK506-BINDING NUCLEAR PROTEIN | | Authors: | Artero, J, Forsyth, T, Callow, P, Watson, A.A, Zhang, W, Laue, E.D, Edlich-Muth, C, Przewloka, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Pentameric Nucleoplasmin Fold is Present in Drosophila Fkbp39 and a Large Number of Chromatin-Related Proteins.
J.Mol.Biol., 427, 2015
|
|
4PBZ
 
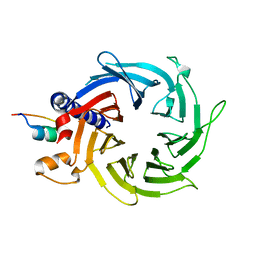 | | Structure of the human RbAp48-MTA1(670-695) complex | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Pei, X.Y, Watson, A.A, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
1BA4
 
 | | THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Coles, M, Bicknell, W, Watson, A.A, Fairlie, D.P, Craik, D.J. | | Deposit date: | 1998-04-07 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is?
Biochemistry, 37, 1998
|
|
1BJC
 
 | | SOLUTION NMR STRUCTURE OF AMYLOID BETA[F16], RESIDUES 1-28, 15 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Poulsen, S.-A, Watson, A.A, Craik, D.J. | | Deposit date: | 1998-06-23 | | Release date: | 1998-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures in aqueous SDS micelles of two amyloid beta peptides of A beta(1-28) mutated at the alpha-secretase cleavage site (K16E, K16F)
J.Struct.Biol., 130, 2000
|
|
1BJB
 
 | | SOLUTION NMR STRUCTURE OF AMYLOID BETA[E16], RESIDUES 1-28, 14 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Poulsen, S.-A, Watson, A.A, Craik, D.J. | | Deposit date: | 1998-06-23 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures in aqueous SDS micelles of two amyloid beta peptides of A beta(1-28) mutated at the alpha-secretase cleavage site (K16E, K16F)
J.Struct.Biol., 130, 2000
|
|
