5VOU
 
 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Chen, S, Zhao, Y, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
5VOT
 
 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Chen, S, Zhao, Y, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
5VOV
 
 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
7WCC
 
 | | Oxidase ChaP-D49L/Q91C mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Sun, M.X, Zheng, W, Wang, Y.S, Zhu, J.P, Tan, R.X. | | Deposit date: | 2021-12-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Oxidase ChaP-D49L/Q91C mutant
To Be Published
|
|
6KL5
 
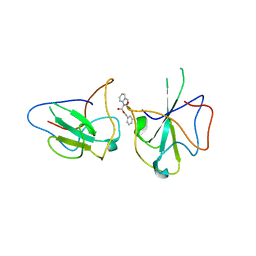 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL6
 
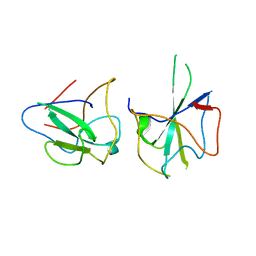 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | Descriptor: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL2
 
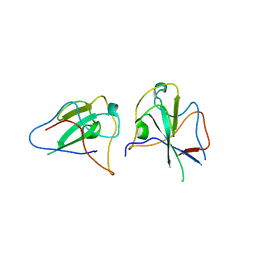 | |
6A4X
 
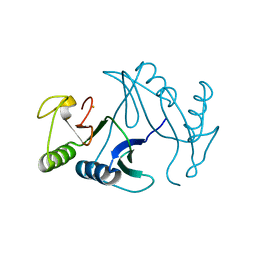 | | Oxidase ChaP-H2 | | Descriptor: | Bleomycin resistance protein, FE (II) ION | | Authors: | Zhang, B, Wang, Y.S, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
