3SV5
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: iodide complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein, ... | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3SSK
 
 | |
3SS0
 
 | |
3SSH
 
 | |
3SSL
 
 | |
3SSY
 
 | |
3SVC
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: chloride complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
6A2W
 
 | | Crystal structure of fucoxanthin chlorophyll a/c complex from Phaeodactylum tricornutum | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W, Yu, L.J, Kuang, T.Y, Shen, J.R. | | Deposit date: | 2018-06-13 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for blue-green light harvesting and energy dissipation in diatoms.
Science, 363, 2019
|
|
3ST0
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: halide-free | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3TAN
 
 | |
3THV
 
 | |
8GYN
 
 | | zebrafish TIPE1 strucutre in complex with PE | | Descriptor: | Tumor necrosis factor alpha-induced protein 8-like protein 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Wang, W, Cao, S.J. | | Deposit date: | 2022-09-23 | | Release date: | 2023-04-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural insight into TIPE1 functioning as a lipid transfer protein.
J.Biomol.Struct.Dyn., 41, 2023
|
|
3TI0
 
 | |
8G7U
 
 | |
8G7T
 
 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-end) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
8G7V
 
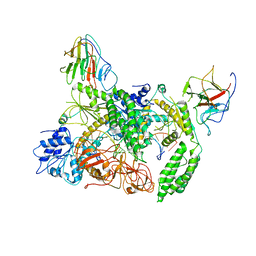 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-inter) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
7BV8
 
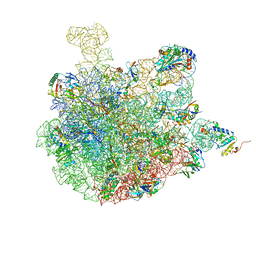 | | Mature 50S ribosomal subunit from RrmJ knock out E.coli strain | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Wang, W, Li, W.Q, Ge, X.L, Yan, K.G, Mandava, C.S, Sanyal, S, Gao, N. | | Deposit date: | 2020-04-09 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Loss of a single methylation in 23S rRNA delays 50S assembly at multiple late stages and impairs translation initiation and elongation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6J0Z
 
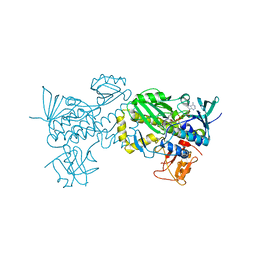 | | Crystal structure of AlpK | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative angucycline-like polyketide oxygenase | | Authors: | Wang, W, Liu, Y, Liang, H. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Crystal structure of AlpK: An essential monooxygenase involved in the biosynthesis of kinamycin
Biochem. Biophys. Res. Commun., 510, 2019
|
|
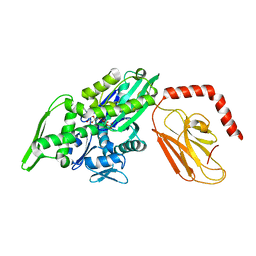
7KRW
 
 | |
7KO2
 
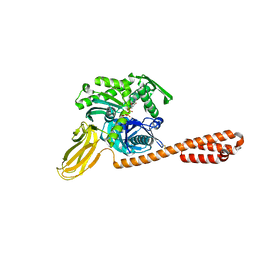 | |
7KRU
 
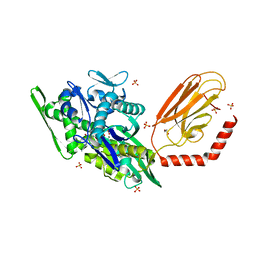 | |
7KRV
 
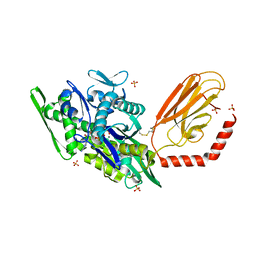 | | Stimulating state of disulfide-bridged Hsp70 DnaK | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK fused with substrate peptide, MAGNESIUM ION, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Conformational equilibria in allosteric control of Hsp70 chaperones.
Mol.Cell, 81, 2021
|
|
7KRT
 
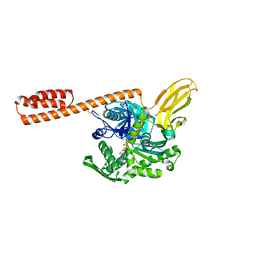 | | Restraining state of a truncated Hsp70 DnaK | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, MAGNESIUM ION | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Conformational equilibria in allosteric control of Hsp70 chaperones.
Mol.Cell, 81, 2021
|
|
7KZI
 
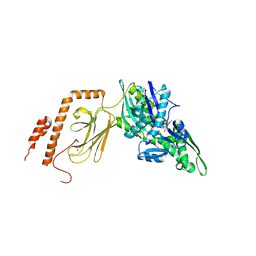 | | Intermediate state (QQQ) of near full-length DnaK alternatively fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7KZU
 
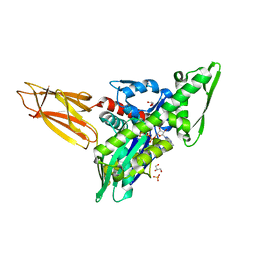 | | Quasi-intermediate state (Q) of a truncated Hsp70 DnaK fused with a substrate peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK fused with substrate peptide,Chaperone protein DnaK fused with substrate peptide, GLYCEROL, ... | | Authors: | Wang, W, Hendrickson, W.A. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intermediates in allosteric equilibria of DnaK-ATP interactions with substrate peptides
Acta Crystallogr.,Sect.D, 77, 2021
|
|
