5CVE
 
 | |
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
4RIZ
 
 | |
4RIT
 
 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-07 | | Release date: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
4RBN
 
 | | The crystal structure of Nitrosomonas europaea sucrose synthase: Insights into the evolutionary origin of sucrose metabolism in prokaryotes | | Descriptor: | Sucrose synthase:Glycosyl transferases group 1 | | Authors: | Wu, R, Asencion Diez, M.D, Figueroa, C.M, Machtey, M, Iglesias, A.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The Crystal Structure of Nitrosomonas europaea Sucrose Synthase Reveals Critical Conformational Changes and Insights into Sucrose Metabolism in Prokaryotes.
J.Bacteriol., 197, 2015
|
|
4RJ0
 
 | |
4RM7
 
 | |
4RUW
 
 | |
4RM1
 
 | |
4RLG
 
 | |
7RI5
 
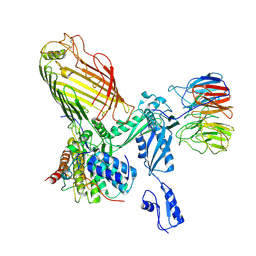 | | Structure of a BAM in MSP1E3D1 nanodiscs at 4 Angstrom resolution | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RJ5
 
 | | The structure of BAM in complex with EspP at 7 Angstrom resolution | | Descriptor: | Maltodextrin-binding protein,Autotransporter outer membrane beta-barrel domain-containing protein chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI6
 
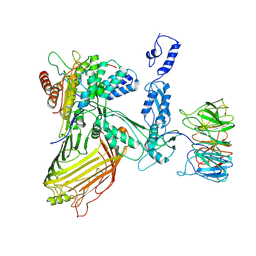 | |
7RI7
 
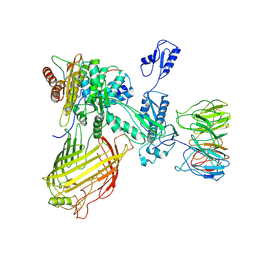 | | The structure of BAM in MSP1D1 nanodiscs | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI4
 
 | | Structure of a BAM/EspP(beta9-12) hybrid-barrel intermediate | | Descriptor: | EspPbeta9-12, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI9
 
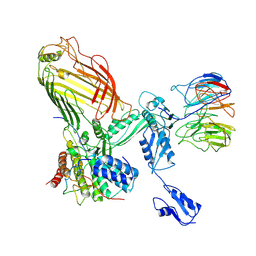 | | The structure of BAM in MSP1E3D1 at 6.9 Angstrom resolution | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI8
 
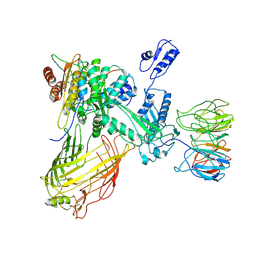 | | The structure of BAM in MSP2N2 nanodiscs | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
4RT5
 
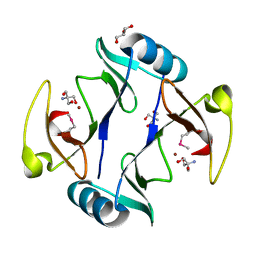 | | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase protein from planctomyces limnophilus dsm 3776 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Wu, R, Bearden, J, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase protein from Planctomyces limnophilus dsm 3776
TO BE PUBLISHED
|
|
4RU0
 
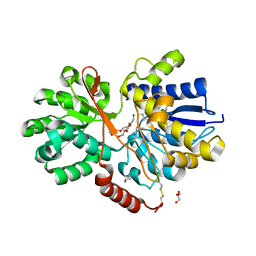 | | The crystal structure of abc transporter permease from pseudomonas fluorescens group | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, GLYCEROL, Putative branched-chain amino acid ABC transporter, ... | | Authors: | Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-17 | | Release date: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | The crystal structure of abc transporter permease from pseudomonas fluorescens group
To be Published
|
|
2I5U
 
 | | Crystal structure of DnaD domain protein from Enterococcus faecalis. Structural genomics target APC85179 | | Descriptor: | DnaD domain protein, MAGNESIUM ION | | Authors: | Wu, R, Zhang, R, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 A crystal structure of a DnaD domain protein from Enterococcus Faecalis
To be Published, 2006
|
|
6UAG
 
 | |
6UG4
 
 | |
6UHH
 
 | | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, R, Kim, Y, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-09-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.138 Å) | | Cite: | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP
To Be Published
|
|
6UAM
 
 | |
1LJ9
 
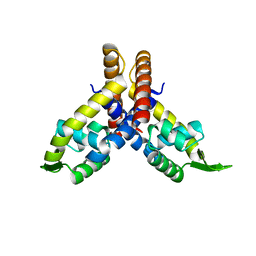 | | The crystal structure of the transcriptional regulator SlyA | | Descriptor: | transcriptional regulator SlyA | | Authors: | Wu, R.Y, Zhang, R.G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-04-19 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor
J.Biol.Chem., 278, 2003
|
|
