3THW
 
 | | Human MutSbeta complexed with an IDL of 4 bases (Loop4) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop4 hairpin, DNA mismatch repair protein Msh2, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
4COD
 
 | | Encoded library technology as a source of hits for the discovery and lead optimization of a potent and selective class of bactericidal direct inhibitors of Mycobacterium tuberculosis InhA | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], N-((3R,5S)-1-(benzofuran-3-carbonyl)-5-(ethylcarbamoyl)pyrrolidin-3-yl)-3-ethyl-1-methyl-1H-pyrazole-5-carboxamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Encinas, L, OKeefe, H, Neu, M, Convery, M.A, McDowell, W, Mendoza-Losana, A, Pages, L.B, Castro-Pichel, J, Evindar, G. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Encoded Library Technology as a Source of Hits for the Discovery and Lead Optimization of a Potent and Selective Class of Bactericidal Direct Inhibitors of Mycobacterium Tuberculosis Inha.
J.Med.Chem., 57, 2014
|
|
4PON
 
 | | The crystal structure of a putative SAM-dependent methyltransferase, YtqB, from Bacillus subtilis | | Descriptor: | Putative RNA methylase | | Authors: | Park, S.C, Song, W.S, Yoon, S.I. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a putative SAM-dependent methyltransferase, YtqB, from Bacillus subtilis
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4CQE
 
 | | B-Raf Kinase V600E mutant in complex with a diarylthiazole B-Raf Inhibitor | | Descriptor: | N-{4-[2-(1-cyclopropylpiperidin-4-yl)-4-(3-{[(2,5-difluorophenyl)sulfonyl]amino}-2-fluorophenyl)-1,3-thiazol-5-yl]pyridin-2-yl}acetamide, SLC45A3-BRAF FUSION PROTEIN | | Authors: | Casale, E, Fasolini, M, Pulici, M, Traquandi, G, Marchionni, C, Modugno, M, Lupi, R, Amboldi, N, Colombo, N, Corti, L, Gasparri, F, Pastori, W, Scolaro, A, Donati, D, Felder, E, Galvani, A, Isacchi, A, Pesenti, E, Ciomei, M. | | Deposit date: | 2014-02-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Optimization of Diarylthiazole B-Raf Inhibitors: Identification of a Compound Endowed with High Oral Antitumor Activity, Mitigated Herg Inhibition, and Low Paradoxical Effect.
Chemmedchem, 10, 2015
|
|
3ST0
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: halide-free | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
4NPM
 
 | |
4NS5
 
 | | Crystal structure of human BS69 Bromo-Zinc finger-PWWP | | Descriptor: | ZINC ION, Zinc finger MYND domain-containing protein 11 | | Authors: | Wang, J.C, Qin, S, Li, F.D, Li, S, Zhang, W, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2013-11-28 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human BS69 Bromo-ZnF-PWWP reveals its role in H3K36me3 nucleosome binding.
Cell Res., 24, 2014
|
|
4CVR
 
 | | Structure of Apobacterioferritin Y25F variant | | Descriptor: | BACTERIOFERRITIN, ZINC ION | | Authors: | Hingorani, K, Pace, R, Whitney, S, Murray, J.W, Wydrzynski, T, Cheah, M.H, Smith, P, Hillier, W. | | Deposit date: | 2014-03-29 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Photo-Oxidation of Tyrosine in a Bio-Engineered Bacterioferritin 'Reaction Centre'-A Protein Model for Artificial Photosynthesis.
Biochim.Biophys.Acta, 1837, 2014
|
|
3U90
 
 | | apoferritin: complex with SDS | | Descriptor: | CADMIUM ION, DODECYL SULFATE, Ferritin light chain | | Authors: | Liu, R, Bu, W, Xi, J, Mortazavi, S.R, Cheung-Lau, J.C, Dmochowski, I.J, Loll, P.J. | | Deposit date: | 2011-10-17 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Beyond the detergent effect: a binding site for sodium dodecyl sulfate (SDS) in mammalian apoferritin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4ND9
 
 | | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation | | Descriptor: | ABC transporter, substrate binding protein (Proline/glycine/betaine) | | Authors: | Nicholls, R, Tkaczuk, K.L, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-25 | | Release date: | 2013-12-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation
To be Published
|
|
1Q6L
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
4PH6
 
 | |
4CZB
 
 | | Structure of the sodium proton antiporter MjNhaP1 from Methanocaldococcus jannaschii at pH 8. | | Descriptor: | NA(+)/H(+) ANTIPORTER 1, POTASSIUM ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Woehlert, D, Paulino, C, Kapotova, E, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 3D Em Map of the Sodium Proton Antiporter Mjnhap1 from Methanocaldococcus Jannaschii
Elife, 3, 2014
|
|
4J77
 
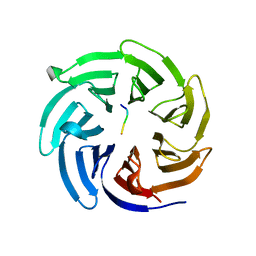 | |
4NWQ
 
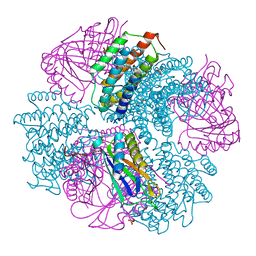 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage, T33-21, Crystallized in Space Group F4132 | | Descriptor: | Putative uncharacterized protein PH0671, SULFATE ION, Uncharacterized protein | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
4JQO
 
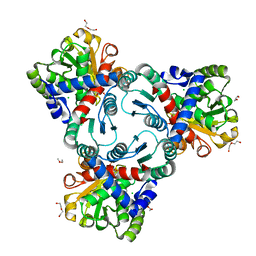 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with citrulline and inorganic phosphate | | Descriptor: | CHLORIDE ION, CITRULLINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Winsor, J, Grimshaw, S, Domagalski, M.J, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
4J9L
 
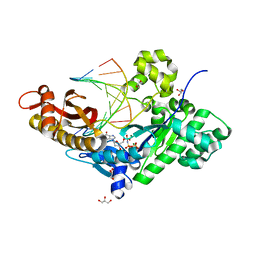 | | Human DNA polymerase eta-DNA ternary complex: misincorporation G opposite T after a C at the primer 3' end (CA/G) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA, DNA polymerase eta, ... | | Authors: | Zhao, Y, Gregory, M, Biertumpfel, C, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2013-02-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of somatic hypermutation at the WA motif by human DNA polymerase eta.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4CXW
 
 | | Crystal structure of human FTO in complex with subfamily-selective inhibitor 12 | | Descriptor: | (2E)-4-[N'-(4-benzyl-pyridine-3-carbonyl)-hydrazino]-4-oxo-but-2-enoic acid, ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE FTO, NICKEL (II) ION | | Authors: | Toh, D.W, Sun, L, Tan, J, Chen, Y, Lau, L.Z.M, Hong, W, Woon, E.C.Y, Gao, Y.G. | | Deposit date: | 2014-04-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor ofN6-methyladenosine demethylase FTO.
Chem Sci, 6, 2015
|
|
4NEK
 
 | | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Geffken, K, Chapman, H.C, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1
To be Published
|
|
3U4A
 
 | | From soil to structure: a novel dimeric family 3-beta-glucosidase isolated from compost using metagenomic analysis | | Descriptor: | CALCIUM ION, JMB19063, beta-D-glucopyranose, ... | | Authors: | McAndrew, R.P, Park, J.I, Reindl, W, Friedland, G.D, D'haeseleer, P, Northen, T, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | From soil to structure: a novel dimeric family 3-beta--glucosidase isolated from compost using metagenomic analysis
To be Published
|
|
4D06
 
 | | Bacterial chalcone isomerase complexed with naringenin | | Descriptor: | (2E)-3-(4-hydroxyphenyl)-1-(2,4,6-trihydroxyphenyl)prop-2-en-1-one, CHALCONE ISOMERASE, CHLORIDE ION, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3TL8
 
 | |
4JUJ
 
 | | Crystal structure of 1918 pandemic influenza virus hemagglutinin mutant D225G complexed with human receptor analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
4NHD
 
 | | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 1, CALCIUM ION, COENZYME A, ... | | Authors: | Hou, J, Zheng, H, Langner, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-04 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A
To be Published
|
|
3THZ
 
 | | Human MutSbeta complexed with an IDL of 6 bases (Loop6) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop6 minus strand, DNA Loop6 plus strand, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Mechanism of mismatch repair revealed by human MutS bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
