3U1L
 
 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
8YHL
 
 | | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoranyl-~{N}-[[5-(6-methylpyridin-2-yl)-4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-1~{H}-imidazol-2-yl]methyl]aniline, ACETATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus
To Be Published
|
|
8YHP
 
 | | Structure of the PGK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Phosphoglycerate kinase 1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the PGK1 from Biortus.
To Be Published
|
|
8YGW
 
 | | The Crystal Structure of MAPK11 from Biortus | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Guo, S. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | The Crystal Structure of MAPK11 from Biortus.
To Be Published
|
|
8YHV
 
 | |
5FKZ
 
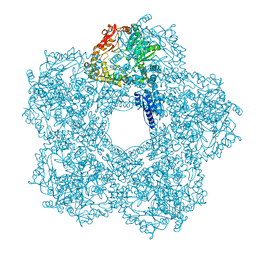 | | Structure of E.coli Constitutive lysine decarboxylase | | Descriptor: | LYSINE DECARBOXYLASE, CONSTITUTIVE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
8XVF
 
 | |
4JYI
 
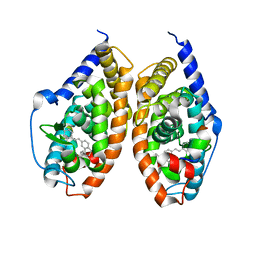 | | Crystal structure of RARbeta LBD in complex with selective partial agonist BMS641 [3-chloro-4-[(E)-2-(5,5-dimethyl-8-phenyl-5,6-dihydronaphthalen-2-yl)ethenyl]benzoic acid] | | Descriptor: | 3-chloro-4-[(E)-2-(5,5-dimethyl-8-phenyl-5,6-dihydronaphthalen-2-yl)ethenyl]benzoic acid, CITRATE ANION, Nuclear receptor coactivator 1, ... | | Authors: | Nadendla, E.K, Teyssier, C, Germain, P, Delfosse, V, Bourguet, W. | | Deposit date: | 2013-03-29 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Unexpected Mode Of Binding Defines BMS948 as A Full Retinoic Acid Receptor beta (RAR beta , NR1B2) Selective Agonist.
Plos One, 10, 2015
|
|
4JYY
 
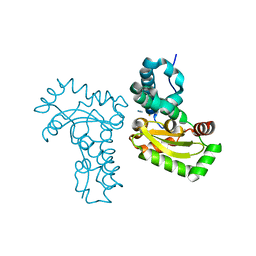 | | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex | | Descriptor: | AZIDE ION, FE (III) ION, Superoxide dismutase | | Authors: | Li, W, Ying, T.L, Wang, C.L, Zhao, Y, Wang, H.F, Tan, X.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structure of the azide and iron substituted Clostrium difficile SOD2 complex
To be Published
|
|
5HFY
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-2-Ala24, beta-3-Lys28, beta-3-Lys31, beta-3-Asn35 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Tavenor, N.A, Reinert, Z.E, Lengyel, G.A, Griffith, B.D, Horne, W.S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold.
Chem.Commun.(Camb.), 52, 2016
|
|
5HG2
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-3-Ala24, beta-3-Lys28, beta-3-Lys31, beta-2-Asn35 | | Descriptor: | GLYCEROL, Immunoglobulin G-binding protein G, MAGNESIUM ION | | Authors: | Tavenor, N.A, Reinert, Z.E, Lengyel, G.A, Griffith, B.D, Horne, W.S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold.
Chem.Commun.(Camb.), 52, 2016
|
|
4JYH
 
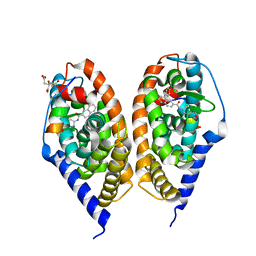 | | Crystal structure of RARbeta LBD in complex with selective agonist BMS948 [4-{[(8-phenylnaphthalen-2-yl)carbonyl]amino}benzoic acid] | | Descriptor: | 4-{[(8-phenylnaphthalen-2-yl)carbonyl]amino}benzoic acid, CITRATE ANION, Nuclear receptor coactivator 1, ... | | Authors: | Nadendla, E.K, Teyssier, C, Germain, P, Delfosse, V, Bourguet, W. | | Deposit date: | 2013-03-29 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Unexpected Mode Of Binding Defines BMS948 as A Full Retinoic Acid Receptor beta (RAR beta , NR1B2) Selective Agonist.
Plos One, 10, 2015
|
|
3VLJ
 
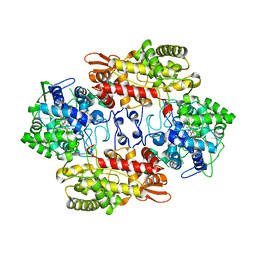 | | Crystal Structure Analysis of the Cyanide Arg409Leu Variant Complexes with o-Dianisidine in KatG from HALOARCULA MARISMORTUI | | Descriptor: | 3,3'-dimethoxybiphenyl-4,4'-diamine, CYANIDE ION, Catalase-peroxidase 2, ... | | Authors: | Sato, T, Higuchi, W, Yoshimatsu, K, Fujiwara, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures and Peroxidatic Function of Cyanide Arg409Leu Variant and its Complexes with o-Dianisidine in KatG from HALOARCULA MARISMORTUI
To be Published
|
|
3VP7
 
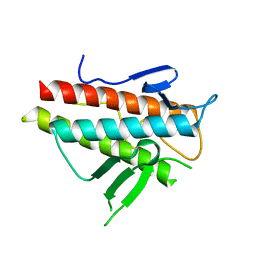 | | Crystal structure of the beta-alpha repeated, autophagy-specific (BARA) domain of Vps30/Atg6 | | Descriptor: | Vacuolar protein sorting-associated protein 30 | | Authors: | Noda, N.N, Kobayashi, T, Adachi, W, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy.
J.Biol.Chem., 287, 2012
|
|
3VPI
 
 | |
8XU5
 
 | |
8XU4
 
 | | The Crystal Structure of MAPK2 from Biortus. | | Descriptor: | MALONIC ACID, MAP kinase-activated protein kinase 2 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Shen, Z. | | Deposit date: | 2024-01-12 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Crystal Structure of MAPK2 from Biortus.
To Be Published
|
|
4JZG
 
 | | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Li, W, Wang, C.L, Zhao, Y, Wang, H.F, Tan, S.X. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Crystal structure of a single cambialistic SOD2 occupied by Manganese ion from Clostridium difficile
To be Published
|
|
3VW5
 
 | |
4JYG
 
 | | Crystal structure of RARbeta LBD in complex with agonist BMS411 [4-{[(5,5-dimethyl-8-phenyl-5,6-dihydronaphthalen-2-yl)carbonyl]amino}benzoic acid] | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-{[(5,5-dimethyl-8-phenyl-5,6-dihydronaphthalen-2-yl)carbonyl]amino}benzoic acid, CITRATE ANION, ... | | Authors: | Nadendla, E.K, Teyssier, C, Germain, P, Delfosse, V, Bourguet, W. | | Deposit date: | 2013-03-29 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An Unexpected Mode Of Binding Defines BMS948 as A Full Retinoic Acid Receptor beta (RAR beta , NR1B2) Selective Agonist.
Plos One, 10, 2015
|
|
8Y4Z
 
 | | Monomeric HERC5 HECT c-lobe structure in solution | | Descriptor: | E3 ISG15--protein ligase HERC5 | | Authors: | Dag, C, Lambert, M, Kahraman, K, Lohn, F, Lee, W, Gocenler, O, Guntert, P, Dotsch, V. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Monomeric HERC5 HECT c-lobe structure in solution
To Be Published
|
|
1HUW
 
 | |
8Y9A
 
 | |
8XX1
 
 | |
4JYJ
 
 | | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Enoyl-CoA hydratase/isomerase, UNKNOWN LIGAND | | Authors: | Cooper, D.R, Porebski, P.J, Domagalski, M.J, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
