6RDW
 
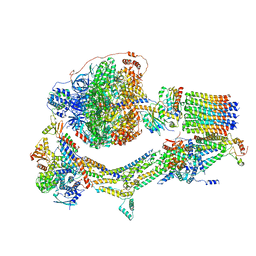 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1F, composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE5
 
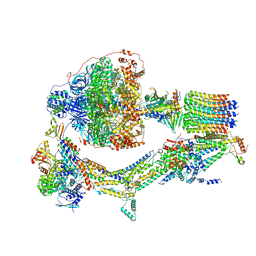 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2C, composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RED
 
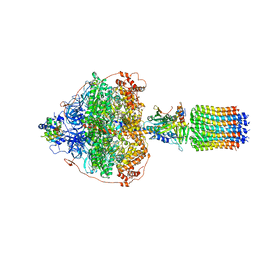 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3A, focussed refinement of F1 head and rotor | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE9
 
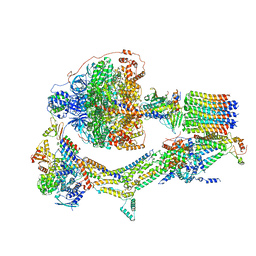 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2D, monomer-masked refinement | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RET
 
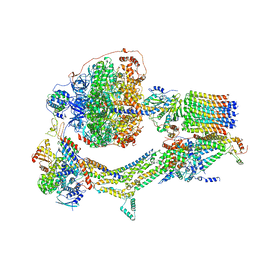 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3C, monomer-masked refinement | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | 著者 | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | 登録日 | 2019-04-12 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
2OLP
 
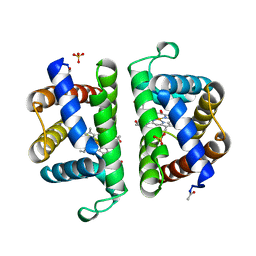 | | Structure and ligand selection of hemoglobin II from Lucina pectinata | | 分子名称: | Hemoglobin II, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Gavira, J.A, Camara-Artigas, A, de Jesus, W, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | 登録日 | 2007-01-19 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.932 Å) | | 主引用文献 | Structure and Ligand Selection of Hemoglobin II from Lucina pectinata
J.Biol.Chem., 283, 2008
|
|
4YJY
 
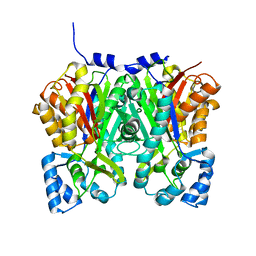 | |
6FKF
 
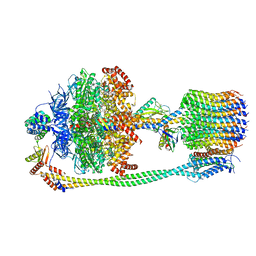 | | Chloroplast F1Fo conformation 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | 著者 | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | 登録日 | 2018-01-24 | | 公開日 | 2018-05-23 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
6IOC
 
 | |
4O61
 
 | | Structure of human ALKBH5 crystallized in the presence of citrate | | 分子名称: | CITRIC ACID, GLYCEROL, RNA demethylase ALKBH5, ... | | 著者 | Tempel, W, Chao, X, Liu, K, Dong, A, Cerovina, T, He, H, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-20 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation.
J.Biol.Chem., 289, 2014
|
|
6IOA
 
 | | The structure of UdgX in complex with uracil | | 分子名称: | IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein, SULFATE ION, ... | | 著者 | Xie, W, Tu, J. | | 登録日 | 2018-10-29 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
3BFD
 
 | |
1ZZS
 
 | | Bovine eNOS N368D single mutant with L-N(omega)-Nitroarginine-(4R)-Amino-L-Proline Amide Bound | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | 著者 | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | 登録日 | 2005-06-14 | | 公開日 | 2005-12-06 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
6YSR
 
 | | Structure of the P+9 stalled ribosome complex | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | 登録日 | 2020-04-23 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
5OQ1
 
 | | Crystal structure of Serratia marcescens ChiX (used as MR model for superior PDB 5OPZ) | | 分子名称: | CHLORIDE ION, ChiX, ZINC ION | | 著者 | Owen, R.A, Fyfe, P.K, Lodge, A, Biboy, J, Vollmer, W, Hunter, W.N, Sargent, F. | | 登録日 | 2017-08-10 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Structure and activity of ChiX: a peptidoglycan hydrolase required for chitinase secretion by Serratia marcescens.
Biochem. J., 475, 2018
|
|
1BOZ
 
 | | STRUCTURE-BASED DESIGN AND SYNTHESIS OF LIPOPHILIC 2,4-DIAMINO-6-SUBSTITUTED QUINAZOLINES AND THEIR EVALUATION AS INHIBITORS OF DIHYDROFOLATE REDUCTASE AND POTENTIAL ANTITUMOR AGENTS | | 分子名称: | N6-(2,5-DIMETHOXY-BENZYL)-N6-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (DIHYDROFOLATE REDUCTASE) | | 著者 | Gangjee, A, Vidwans, A.P, Vasudevan, A, Queener, S.F, Kisliuk, R.L, Cody, V, Li, R, Galitsky, N, Luft, J.R, Pangborn, W. | | 登録日 | 1998-08-06 | | 公開日 | 1998-08-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation as inhibitors of dihydrofolate reductases and potential antitumor agents.
J.Med.Chem., 41, 1998
|
|
5OW7
 
 | | VDR complex | | 分子名称: | (3~{S})-3-[(1~{S},3~{a}~{S},4~{E},7~{a}~{S})-7~{a}-methyl-4-[(2~{Z})-2-[(5~{S})-2-methylidene-5-oxidanyl-cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-3-oxidanyl-~{N}-propan-2-yl-butanamide, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Rochel, N, Li, W. | | 登録日 | 2017-08-31 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
1BS7
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM | | 分子名称: | NICKEL (II) ION, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
3ARQ
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with IDARUBICIN | | 分子名称: | Chitinase A, GLYCEROL, IDARUBICIN | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3AS3
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | 分子名称: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
8IDE
 
 | | Structure of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme (TsaN) | | 分子名称: | MANGANESE (II) ION, N(6)-L-threonylcarbamoyladenine synthase | | 著者 | Zhang, Z.L, Jin, M.Q, Yu, Z.J, Chen, W, Wang, X.L, Lei, D.S, Zhang, W.H. | | 登録日 | 2023-02-13 | | 公開日 | 2023-07-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Structure-function analysis of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme reveals a prototype of tRNA t6A and ct6A synthetases.
Nucleic Acids Res., 51, 2023
|
|
4N7R
 
 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | 分子名称: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | 著者 | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | 登録日 | 2013-10-16 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ARZ
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | 分子名称: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | 著者 | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | 登録日 | 2010-12-09 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
6TAH
 
 | |
6TH5
 
 | |
