6DPP
 
 | |
1G2A
 
 | | THE CRYSTAL STRUCTURE OF E.COLI PEPTIDE DEFORMYLASE COMPLEXED WITH ACTINONIN | | Descriptor: | ACTINONIN, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-18 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
3NII
 
 | | The structure of UBR box (KIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide KIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
1MRB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RABBIT LIVER CD7 METALLOTHIONEIN-2A IN AQUEOUS SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2A | | Authors: | Braun, W, Arseniev, A, Schultze, P, Woergoetter, E, Wagner, G, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance.
J.Mol.Biol., 201, 1988
|
|
2OWY
 
 | |
3RGZ
 
 | | Structural insight into brassinosteroid perception by BRI1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chai, J, Han, Z, She, J, Wang, J, Cheng, W. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural insight into brassinosteroid perception by BRI1.
Nature, 474, 2011
|
|
2X3V
 
 | | Structure of The F-BAR Domain of Mouse Syndapin I | | Descriptor: | PROTEIN KINASE C AND CASEIN KINASE SUBSTRATE IN NEURONS PROTEIN 1 | | Authors: | Ma, Q, Rao, Y, Vahedi-Faridi, A, Saenger, W, Haucke, V. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular Basis for SH3 Domain Regulation of F-Bar-Mediated Membrane Deformation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NCP
 
 | | GlnK2 from Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, Nitrogen regulatory protein P-II (GlnB-2) | | Authors: | Helfmann, S, Lue, W, Litz, C, Andrade, S.L.A. | | Deposit date: | 2010-06-05 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cooperative binding of MgATP and MgADP in the trimeric P(II) protein GlnK2 from Archaeoglobus fulgidus.
J.Mol.Biol., 402, 2010
|
|
2XAT
 
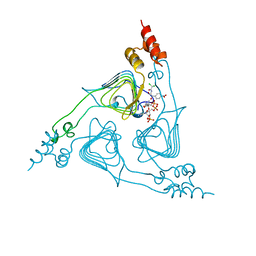 | |
3N78
 
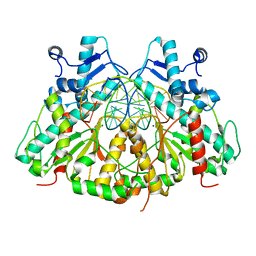 | | SgrAI bound to Secondary Site DNA and Mg(II) | | Descriptor: | DNA (5'-D(*AP*GP*TP*CP*CP*AP*CP*CP*GP*GP*GP*GP*GP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*CP*CP*CP*CP*CP*GP*GP*TP*GP*GP*AP*CP*T)-3'), MAGNESIUM ION, ... | | Authors: | Horton, N.C, Little, E.J, Dunten, P.W. | | Deposit date: | 2010-05-26 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | New clues in the allosteric activation of DNA cleavage by SgrAI: structures of SgrAI bound to cleaved primary-site DNA and uncleaved secondary-site DNA.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1E8F
 
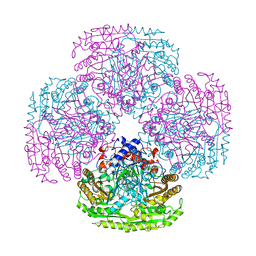 | |
2WNJ
 
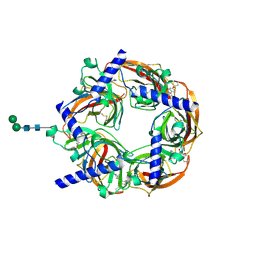 | | CRYSTAL STRUCTURE OF APLYSIA ACHBP IN COMPLEX WITH DMXBA | | Descriptor: | (3E)-3-[(2,4-DIMETHOXYPHENYL)METHYLIDENE]-3,4,5,6-TETRAHYDRO-2,3'-BIPYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-09 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Determinants for Interaction of Partial Agonists with Acetylcholine Binding Protein and Neuronal Alpha7 Nicotinic Acetylcholine Receptor.
Embo J., 28, 2009
|
|
2WSW
 
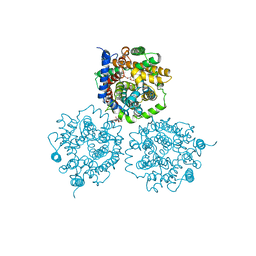 | | Crystal Structure of Carnitine Transporter from Proteus mirabilis | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, BCCT FAMILY BETAINE/CARNITINE/CHOLINE TRANSPORTER, GLYCEROL, ... | | Authors: | Schulze, S, Terwisscha van Scheltinga, A.C, Kuehlbrandt, W. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural Basis of Na(+)-Independent and Cooperative Substrate/Product Antiport in Cait.
Nature, 467, 2010
|
|
1NWI
 
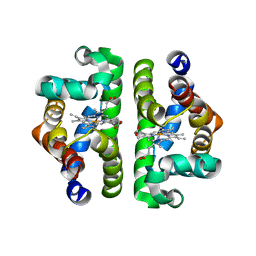 | |
1EUC
 
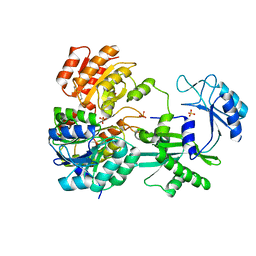 | | CRYSTAL STRUCTURE OF DEPHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE | | Descriptor: | PHOSPHATE ION, SUCCINYL-COA SYNTHETASE, ALPHA CHAIN, ... | | Authors: | Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 2000-04-14 | | Release date: | 2000-07-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA synthetase.
J.Mol.Biol., 299, 2000
|
|
3N9Y
 
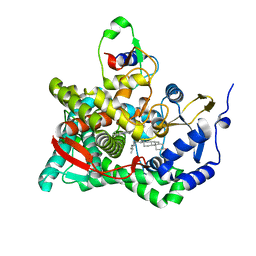 | | Crystal structure of human CYP11A1 in complex with cholesterol | | Descriptor: | Adrenodoxin, CHOLESTEROL, Cholesterol side-chain cleavage enzyme, ... | | Authors: | Strushkevich, N.V, MacKenzie, F, Tempel, W, Botchkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J.U, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2WYQ
 
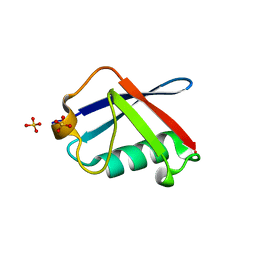 | |
3Q6J
 
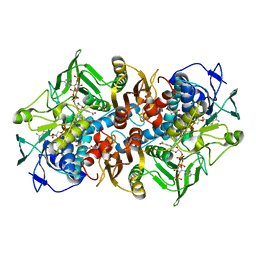 | | Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M Oxidoreductase/Carboxylase | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, ... | | Authors: | Pandey, A.S, Mulder, D.W, Ensign, S.A, Peters, J.W. | | Deposit date: | 2011-01-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M oxidoreductase/carboxylase.
Febs Lett., 585, 2011
|
|
1E6X
 
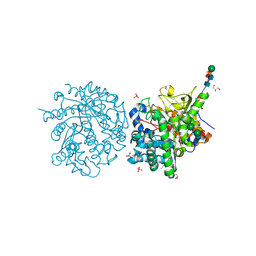 | | MYROSINASE FROM SINAPIS ALBA with a bound transition state analogue,D-glucono-1,5-lactone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-glucono-1,5-lactone, ... | | Authors: | Burmeister, W.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Resolution X-Ray Crystallography Shows that Ascorbate is a Cofactor for Myrosinase and Substitutes for the Function of the Catalytic Base
J.Biol.Chem., 275, 2000
|
|
4DFP
 
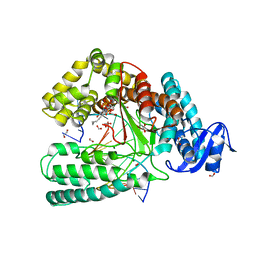 | | Crystal structure of the large fragment of DNA Polymerase I from Thermus aqauticus in a ternary complex with 7-(aminopentinyl)-7-deaza-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-5-(5-aminopent-1-yn-1-yl)-7-{2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 5'-d(AAACGGCGCCGTGGTC)-3', ... | | Authors: | Bergen, K, Steck, A, Struett, S, Baccaro, A, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2012-01-24 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of KlenTaq DNA Polymerase Caught While Incorporating C5-Modified Pyrimidine and C7-Modified 7-Deazapurine Nucleoside Triphosphates.
J.Am.Chem.Soc., 134, 2012
|
|
6E1X
 
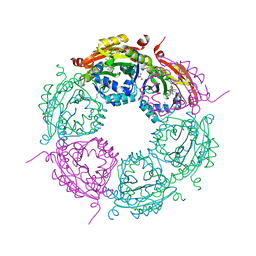 | | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-10 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To Be Published
|
|
1EXY
 
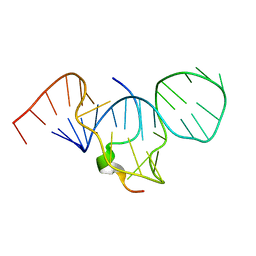 | | SOLUTION STRUCTURE OF HTLV-1 PEPTIDE BOUND TO ITS RNA APTAMER TARGET | | Descriptor: | HTLV-1 REX PEPTIDE, RNA APTAMER, 33-MER | | Authors: | Jiang, F, Gorin, A, Hu, W, Majumdar, A, Baskerville, S, Xu, W, Ellington, A, Patel, D.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples.
Structure Fold.Des., 7, 1999
|
|
4DKR
 
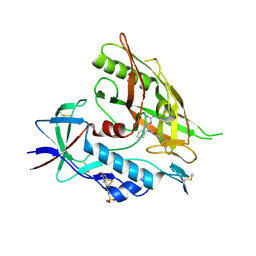 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with AWS-I-169 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
4DDF
 
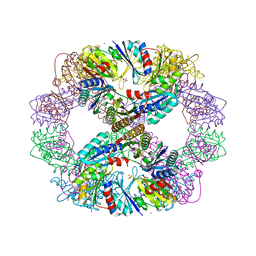 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
3NIJ
 
 | | The structure of UBR box (HIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
