3HZ3
 
 | | Lactobacillus reuteri N-terminally truncated glucansucrase GTF180(D1025N)-sucrose complex | | Descriptor: | CALCIUM ION, Glucansucrase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-06-23 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2GVY
 
 | |
3KLK
 
 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in triclinic apo- form | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KLL
 
 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180-maltose complex | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase, ... | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3DTK
 
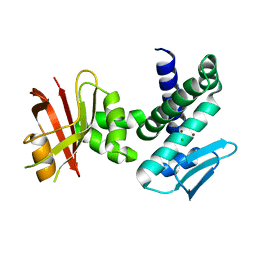 | | Crystal structure of the IRRE protein, a central regulator of DNA damage repair in deinococcaceae | | Descriptor: | CHLORIDE ION, IRRE PROTEIN, ZINC ION | | Authors: | Vujicic-Zagar, A, Dulermo, R, Le Gorrec, M, Vannier, F, Servant, P, Sommer, S, de Groot, A, Serre, L. | | Deposit date: | 2008-07-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Crystal structure of the IrrE protein, a central regulator of DNA damage repair in deinococcaceae
J.Mol.Biol., 386, 2009
|
|
3DTI
 
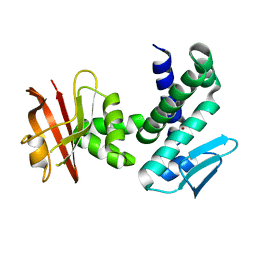 | | Crystal structure of the IRRE protein, a central regulator of DNA damage repair in deinococcaceae | | Descriptor: | IRRE protein, ZINC ION | | Authors: | Vujicic-Zagar, A, Dulermo, R, Le Gorrec, M, Vannier, F, Servant, P, Sommer, S, De Groot, A, Serre, L. | | Deposit date: | 2008-07-15 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the IrrE protein, a central regulator of DNA damage repair in deinococcaceae
J.Mol.Biol., 386, 2009
|
|
3DTE
 
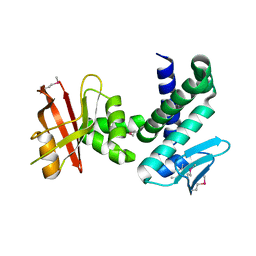 | | Crystal structure of the IRRE protein, a central regulator of DNA damage repair in deinococcaceae | | Descriptor: | IrrE protein | | Authors: | Vujicic-Zagar, A, Dulermo, R, Le Gorrec, M, Vannier, F, Servant, P, Sommer, S, De Groot, A, Serre, L. | | Deposit date: | 2008-07-15 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the IrrE protein, a central regulator of DNA damage repair in deinococcaceae
J.Mol.Biol., 386, 2009
|
|
2GUY
 
 | |
4KQP
 
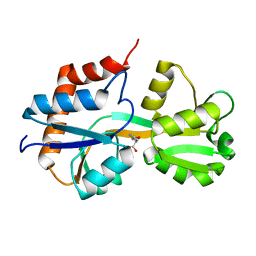 | | Crystal structure of Lactococcus lactis GlnP substrate binding domain 2 (SBD2) in complex with glutamine at 0.95 A resolution | | Descriptor: | GLUTAMINE, Glutamine ABC transporter permease and substrate binding protein protein | | Authors: | Vujicic Zagar, A, Guskov, A, Schuurman-Wolters, G.K, Slotboom, D.J, Poolman, B. | | Deposit date: | 2013-05-15 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
4KPT
 
 | | Crystal structure of substrate binding domain 1 (SBD1) OF ABC transporter GLNPQ from lactococcus lactis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vujicic Zagar, A, Guskov, A, Schuurman-Wolters, G.K, Slotboom, D.J, Poolman, B. | | Deposit date: | 2013-05-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
4KR5
 
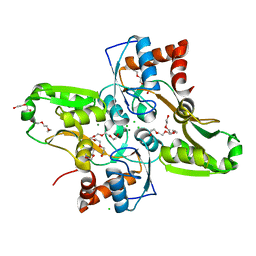 | | Crystal structure of Lactococcus lactis GlnP substrate binding domain 2 (SBD2) in open conformation | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Vujicic Zagar, A, Guskov, A, Schuurman-Wolters, G.K, Slotboom, D.J, Poolman, B. | | Deposit date: | 2013-05-16 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
8B57
 
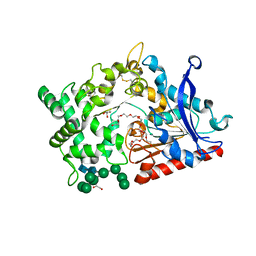 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-09-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
8BBX
 
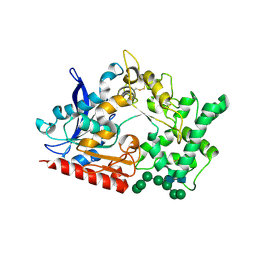 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 in space group C222(1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoprotease endo-Pro, TETRAETHYLENE GLYCOL, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
4AYG
 
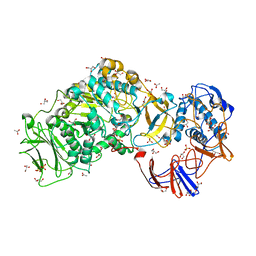 | | Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in orthorhombic apo-form | | Descriptor: | ACETIC ACID, CALCIUM ION, GLUCANSUCRASE, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2012-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of Truncated and Full-Length Glucansucrase Gtf180 Enzymes from Lactobacillus Reuteri 180.
FEBS J., 281, 2014
|
|
4AMC
 
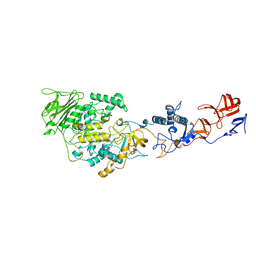 | | Crystal structure of Lactobacillus reuteri 121 N-terminally truncated glucansucrase GTFA | | Descriptor: | CALCIUM ION, GLUCANSUCRASE | | Authors: | Pijning, T, Vujicic-Zagar, A, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2012-03-08 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the Alpha-1,6/Alpha-1,4-Specific Glucansucrase Gtfa from Lactobacillus Reuteri 121
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4C1T
 
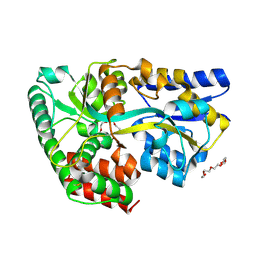 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with arabinoxylotriose | | Descriptor: | PENTAETHYLENE GLYCOL, SUGAR TRANSPORTER SOLUTE-BINDING PROTEIN, alpha-L-arabinofuranose-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fredslund, F, Ejby, M, Vujicic-Zagar, A, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
4C1U
 
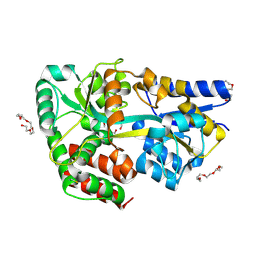 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with arabinoxylobiose | | Descriptor: | PENTAETHYLENE GLYCOL, SUGAR TRANSPORTER SOLUTE-BINDING PROTEIN, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fredslund, F, Ejby, M, Vujicic-Zagar, A, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
3ZKL
 
 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with xylotriose | | Descriptor: | PENTAETHYLENE GLYCOL, PUTATIVE SUGAR TRANSPORTER SOLUTE-BINDING PROTEIN, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Ejby, M, Vujicic-Zagar, A, Fredslund, F, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
3ZKK
 
 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with xylotetraose | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, XOS BINDING PROTEIN, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Ejby, M, Vujicic-Zagar, A, Fredslund, F, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
3RLB
 
 | | Crystal structure at 2.0 A of the S-component for thiamin from an ECF-type ABC transporter | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ThiT, nonyl beta-D-glucopyranoside | | Authors: | Erkens, G.B, Berntsson, R.P.-A, Fulyani, F, Majsnerowska, M, Vujicic-Zagar, A, ter Beek, J, Poolman, B, Slotboom, D.J. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of modularity in ECF-type ABC transporters.
Nat.Struct.Mol.Biol., 18, 2011
|
|
