5ITP
 
 | |
5ITO
 
 | |
6EQ0
 
 | |
6EPZ
 
 | |
6EQ1
 
 | |
6EQ8
 
 | |
6FT2
 
 | |
5HDJ
 
 | | Structure of B. megaterium NfrA1 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NfrA1 | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2016-01-05 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Functional and structural characterization of two Bacillus megaterium nitroreductases biotransforming the herbicide mesotrione.
Biochem.J., 473, 2016
|
|
5HEI
 
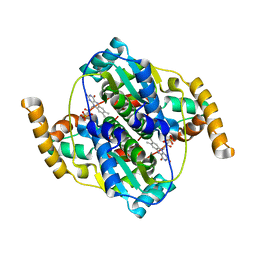 | | Structure of B. megaterium NfrA2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN MONONUCLEOTIDE, NfrA2, ... | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2016-01-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Functional and structural characterization of two Bacillus megaterium nitroreductases biotransforming the herbicide mesotrione.
Biochem.J., 473, 2016
|
|
5OTA
 
 | |
5ORE
 
 | |
5OT8
 
 | |
5OT9
 
 | |
5OTC
 
 | |
5OVZ
 
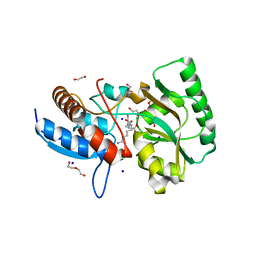 | | High resolution structure of the PBP NocT in complex with nopaline | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(1S)-4-carbamimidamido-1-carboxybutyl]-D-glutamic acid, ... | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2017-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
5ORG
 
 | |
4P0I
 
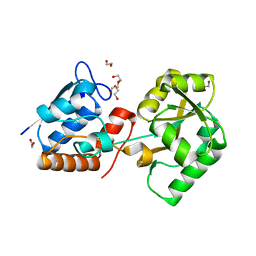 | | Structure of the PBP NocT | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Nopaline-binding periplasmic protein | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
6ZK3
 
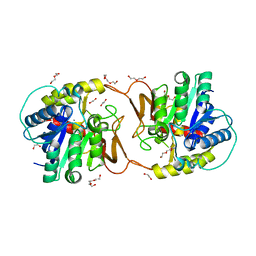 | | Plant nucleoside hydrolase - ZmNRh2b in complex with ribose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK4
 
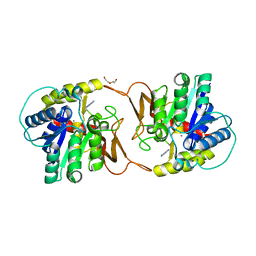 | | Plant nucleoside hydrolase - ZmNRh2b with a bound adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK5
 
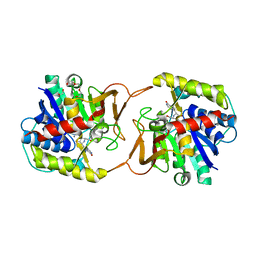 | | Plant nucleoside hydrolase - ZmNRh3 enzyme in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK1
 
 | | Plant nucleoside hydrolase - ZmNRh2b enzyme | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK2
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with forodesine | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2020-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
1WYY
 
 | | Post-fusion hairpin conformation of the sars coronavirus spike glycoprotein | | Descriptor: | CHLORIDE ION, E2 Glycoprotein | | Authors: | Duquerroy, S, Vigouroux, A, Rottier, P.J.M, Rey, F.A, Bosch, B.J. | | Deposit date: | 2005-02-18 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein
Virology, 335, 2005
|
|
1RER
 
 | | Crystal structure of the homotrimer of fusion glycoprotein E1 from Semliki Forest Virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gibbons, D.L, Vaney, M.C, Roussel, A, Vigouroux, A, Reilly, B, Kielian, M, Rey, F.A. | | Deposit date: | 2003-11-07 | | Release date: | 2004-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus.
Nature, 427, 2004
|
|
6EPY
 
 | |
