3NT4
 
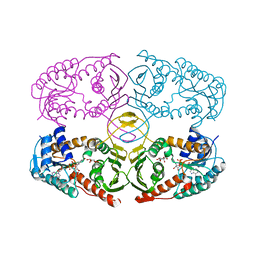 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADH and inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NTQ
 
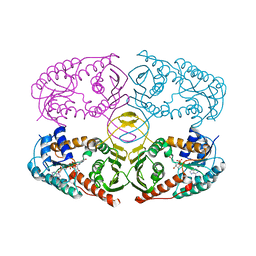 | |
3NTR
 
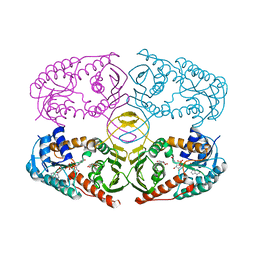 | | Crystal structure of K97V mutant of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD and inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6503 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
6DEH
 
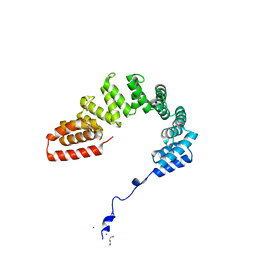 | | Structure of LpnE Effector Protein from Legionella pneumophila (sp. Philadelphia) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Voth, K, Chung, I.Y.W, van Straaten, K.E, Cygler, M. | | Deposit date: | 2018-05-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Legionella effector protein LpnE provides insights into its interaction with Oculocerebrorenal syndrome of Lowe (OCRL) protein.
FEBS J., 286, 2019
|
|
3HDQ
 
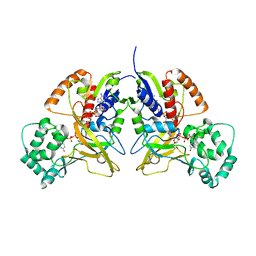 | |
3HE3
 
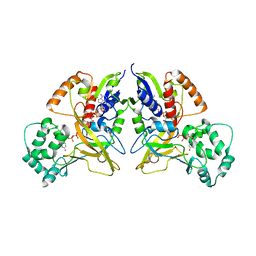 | |
3HDY
 
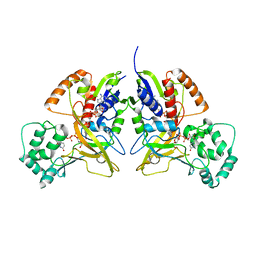 | | Crystal Structure of UDP-galactopyranose mutase (reduced form) in complex with substrate | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Partha, S.K, van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of substrate binding to UDP-galactopyranose mutase: crystal structures in the reduced and oxidized state complexed with UDP-galactopyranose and UDP.
J.Mol.Biol., 394, 2009
|
|
