1MHO
 
 | | THE 2.0 A STRUCTURE OF HOLO S100B FROM BOVINE BRAIN | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Matsumura, H, Shiba, T, Inoue, T, Harada, S, Yasushi, K.A.I. | | Deposit date: | 1997-09-11 | | Release date: | 1998-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain.
Structure, 6, 1998
|
|
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4LCD
 
 | |
2CW8
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, GLYCEROL, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
3WRX
 
 | | Crystal structure of helicase complex 1 | | Descriptor: | CESIUM ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8GZV
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZX
 
 | | Escherichia coli FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZW
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZY
 
 | | Escherichia coli FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
4YSV
 
 | |
3VQR
 
 | | Structure of a dye-linked L-proline dehydrogenase mutant from the aerobic hyperthermophilic archaeon, Aeropyrum pernix | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sakuraba, H, Ohshima, T, Satomura, T, Yoneda, K. | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Novel Dye-linked L-Proline Dehydrogenase from Hyperthermophilic Archaeon Aeropyrum pernix
J.Biol.Chem., 287, 2012
|
|
1K1X
 
 | | Crystal structure of 4-alpha-glucanotransferase from thermococcus litoralis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
1K1W
 
 | | Crystal structure of 4-alpha-glucanotransferase from thermococcus litoralis | | Descriptor: | 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
1JQN
 
 | |
1K1Y
 
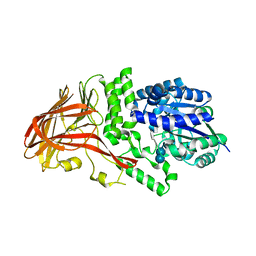 | | Crystal structure of thermococcus litoralis 4-alpha-glucanotransferase complexed with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-ALPHA-GLUCANOTRANSFERASE, ... | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
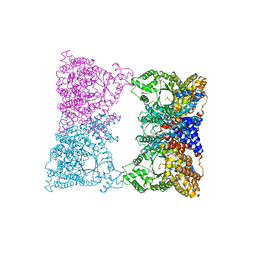
1JQO
 
 | |
2YVK
 
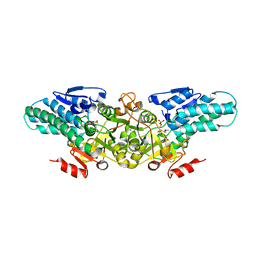 | | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis | | Descriptor: | 5-S-METHYL-1-O-PHOSPHONO-5-THIO-D-RIBULOSE, Methylthioribose-1-phosphate isomerase | | Authors: | Tamura, H, Inoue, T, Kai, Y, Matsumura, H. | | Deposit date: | 2007-04-13 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: Implications for catalytic mechanism
Protein Sci., 17, 2008
|
|
8HYE
 
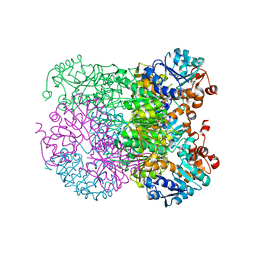 | | Structure of amino acid dehydrogenase-2752 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine dehydrogenase, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
8HYH
 
 | | Structure of amino acid dehydrogenase3448 | | Descriptor: | 1,2-ETHANEDIOL, Alanine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Two different alanine dehydrogenases from Geobacillus kaustophilus: Their biochemical characteristics and differential expression in vegetative cells and spores.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
8J1G
 
 | | Structure of amino acid dehydrogenase in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | First crystal structure of an NADP + -dependent l-arginine dehydrogenase belonging to the mu-crystallin family.
Int.J.Biol.Macromol., 249, 2023
|
|
8J1C
 
 | |
2ZVI
 
 | | Crystal structure of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis | | Descriptor: | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase | | Authors: | Tamura, H, Yadani, T, Kai, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2008-11-07 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the apo decarbamylated form of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2YRF
 
 | | Crystal structure of 5-methylthioribose 1-phosphate isomerase from Bacillus subtilis complexed with sulfate ion | | Descriptor: | Methylthioribose-1-phosphate isomerase, SULFATE ION | | Authors: | Tamura, H, Inoue, T, Kai, Y, Matsumura, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: Implications for catalytic mechanism
Protein Sci., 17, 2008
|
|
1VCV
 
 | |
1WZU
 
 | | Crystal structure of quinolinate synthase (nadA) | | Descriptor: | D-MALATE, Quinolinate synthetase A | | Authors: | Sakuraba, H. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the NAD Biosynthetic Enzyme Quinolinate Synthase
J.Biol.Chem., 280, 2005
|
|
