3VNQ
 
 | |
1X0K
 
 | | Crystal Structure of Bacteriorhodopsin at pH 10 | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Okumura, H, Murakami, M, Kouyama, T. | | Deposit date: | 2005-03-23 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Acid Blue and Alkaline Purple Forms of Bacteriorhodopsin
J.Mol.Biol., 351, 2005
|
|
3VNR
 
 | |
1X0I
 
 | | Crystal Structure of the Acid Blue Form of Bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Okumura, H, Murakami, M, Kouyama, T. | | Deposit date: | 2005-03-23 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Acid Blue and Alkaline Purple Forms of Bacteriorhodopsin
J.Mol.Biol., 351, 2005
|
|
6J84
 
 | | Crystal structure of TleB with hydroxyl analog | | Descriptor: | (2S)-2-hydroxy-N-[(2S)-1-hydroxy-3-(1H-indol-3-yl)propan-2-yl]-3-methylbutanamide, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakamura, H, Mori, T, Abe, I. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
4XB2
 
 | | Hyperthermophilic archaeal homoserine dehydrogenase mutant in complex with NADPH | | Descriptor: | 319aa long hypothetical homoserine dehydrogenase, L-HOMOSERINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
4XB1
 
 | | Hyperthermophilic archaeal homoserine dehydrogenase in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 319aa long hypothetical homoserine dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
5B3F
 
 | |
3WB1
 
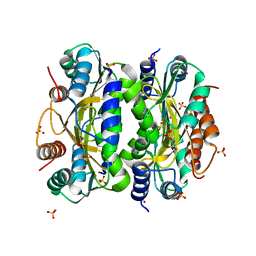 | | HcgB from Methanocaldococcus jannaschii | | Descriptor: | SULFATE ION, Uncharacterized protein MJ0488 | | Authors: | Tamura, H, Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2013-05-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the HcgB enzyme in [Fe]-hydrogenase-cofactor biosynthesis.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3WRY
 
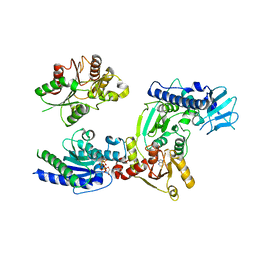 | | Crystal structure of helicase complex 2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WRX
 
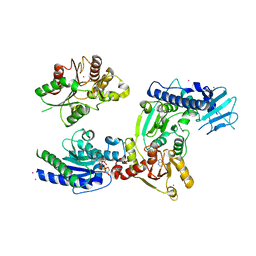 | | Crystal structure of helicase complex 1 | | Descriptor: | CESIUM ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4JJG
 
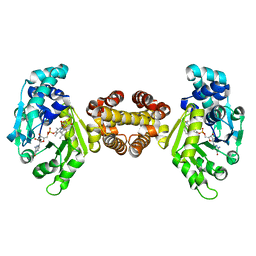 | | Crystal structure of FE-hydrogenase from methanothermobacter marburgensis in complex with toluenesulfonylmethylisocyanide | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase, N-methyl-1-[(4-methylbenzyl)sulfonyl]methanamine, iron-guanylyl pyridinol cofactor | | Authors: | Tamura, H, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2013-03-07 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of [fe]-hydrogenase in complex with inhibitory isocyanides: implications for the h2 -activation site.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3PHF
 
 | |
7VO1
 
 | | Structure of aminotransferase-substrate complex | | Descriptor: | 454aa long hypothetical 4-aminobutyrate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Sakuraba, H, Ohshida, T, Ohshima, T. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of a novel type of ornithine delta-aminotransferase from the hyperthermophilic archaeon Pyrococcus horikoshii.
Int.J.Biol.Macromol., 208, 2022
|
|
4JJF
 
 | | Crystal structure of FE-hydrogenase from methanothermobacter marburgensis in complex with 2-naphthylisocyanide | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase, N-(naphthalen-2-yl)methanimine, iron-guanylyl pyridinol cofactor | | Authors: | Tamura, H, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2013-03-07 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of [fe]-hydrogenase in complex with inhibitory isocyanides: implications for the h2 -activation site.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2YVK
 
 | | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis | | Descriptor: | 5-S-METHYL-1-O-PHOSPHONO-5-THIO-D-RIBULOSE, Methylthioribose-1-phosphate isomerase | | Authors: | Tamura, H, Inoue, T, Kai, Y, Matsumura, H. | | Deposit date: | 2007-04-13 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: Implications for catalytic mechanism
Protein Sci., 17, 2008
|
|
3VTF
 
 | | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3WRV
 
 | |
4YSV
 
 | |
2ZVI
 
 | | Crystal structure of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis | | Descriptor: | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase | | Authors: | Tamura, H, Yadani, T, Kai, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2008-11-07 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the apo decarbamylated form of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1QB4
 
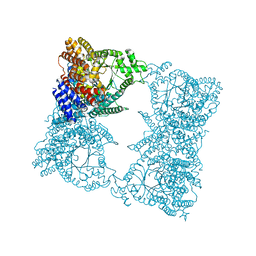 | | CRYSTAL STRUCTURE OF MN(2+)-BOUND PHOSPHOENOLPYRUVATE CARBOXYLASE | | Descriptor: | ASPARTIC ACID, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Matsumura, H, Terada, M, Shirakata, S, Inoue, T, Yoshinaga, T, Izui, K, Kai, Y. | | Deposit date: | 1999-04-30 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plausible phosphoenolpyruvate binding site revealed by 2.6 A structure of Mn2+-bound phosphoenolpyruvate carboxylase from Escherichia coli
FEBS Lett., 458, 1999
|
|
1MHO
 
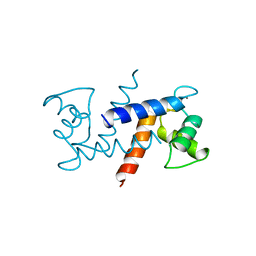 | | THE 2.0 A STRUCTURE OF HOLO S100B FROM BOVINE BRAIN | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Matsumura, H, Shiba, T, Inoue, T, Harada, S, Yasushi, K.A.I. | | Deposit date: | 1997-09-11 | | Release date: | 1998-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain.
Structure, 6, 1998
|
|
2YRF
 
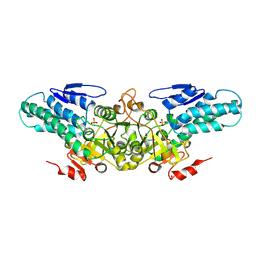 | | Crystal structure of 5-methylthioribose 1-phosphate isomerase from Bacillus subtilis complexed with sulfate ion | | Descriptor: | Methylthioribose-1-phosphate isomerase, SULFATE ION | | Authors: | Tamura, H, Inoue, T, Kai, Y, Matsumura, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: Implications for catalytic mechanism
Protein Sci., 17, 2008
|
|
3STZ
 
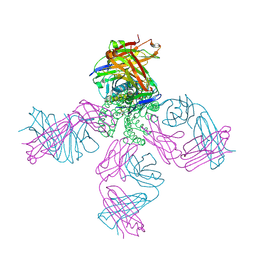 | | KcsA potassium channel mutant Y82C with nitroxide spin label | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
