8CDA
 
 | | Crystal structure of MAB_4123 from Mycobacterium abscessus | | Descriptor: | GLYCEROL, PHOSPHATE ION, Probable monooxygenase | | Authors: | Ung, K.L, Poussineau, C, Couston, J, Alsarraf, H.M.A.B, Blaise, M. | | Deposit date: | 2023-01-30 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MAB_4123, a putative flavin-dependent monooxygenase from Mycobacterium abscessus.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7QPA
 
 | | Outward-facing auxin bound form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
7QP9
 
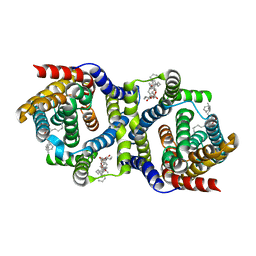 | | Outward-facing apo-form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
7QPC
 
 | | Inward-facing NPA bound form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
6YCA
 
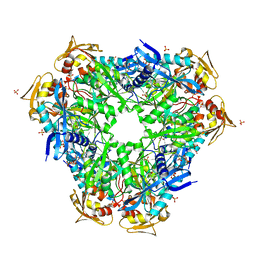 | | Crystal structure of Eis1 from Mycobacterium abscessus | | Descriptor: | ACETYL COENZYME *A, SULFATE ION, Uncharacterized N-acetyltransferase D2E76_00625 | | Authors: | Blaise, M, Ung, K.L. | | Deposit date: | 2020-03-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the N-acetyltransferase Eis1 from Mycobacterium abscessus reveals the molecular determinants of its incapacity to modify aminoglycosides.
Proteins, 89, 2021
|
|
6RFT
 
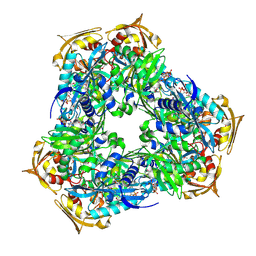 | | Crystal structure of Eis2 from Mycobacterium abscessus bound to Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Uncharacterized N-acetyltransferase D2E36_21790 | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6RFX
 
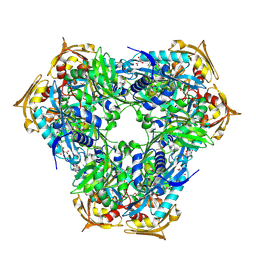 | | Crystal structure of Eis2 from Mycobacterium abscessus | | Descriptor: | ACETATE ION, CITRIC ACID, Eis2, ... | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6RFY
 
 | | Crystal structure of Eis2 form Mycobacterium abscessus | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Eis2, SULFATE ION | | Authors: | Blaise, M, Kremer, L, Olieric, V, Alsarraf, H, Ung, K.L. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the aminoglycosides N-acetyltransferase Eis2 from Mycobacterium abscessus.
Febs J., 286, 2019
|
|
6MEA
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate ribose (ADP-ribose) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6MEN
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6MEB
 
 | | Crystal structure of Tylonycteris bat coronavirus HKU4 macrodomain in complex with nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
2H80
 
 | | NMR structures of SAM domain of Deleted in Liver Cancer 2 (DLC2) | | Descriptor: | StAR-related lipid transfer protein 13 | | Authors: | Li, H.Y, Fung, K.L, Jin, D.Y, Chung, S.S, Ko, B.C, Sun, H.Z. | | Deposit date: | 2006-06-06 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2
Proteins, 67, 2007
|
|
