2CJH
 
 | |
2CJD
 
 | |
2CJG
 
 | | Lysine aminotransferase from M. tuberculosis in bound PMP form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-LYSINE-EPSILON AMINOTRANSFERASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2006-04-01 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Direct Evidence for a Glutamate Switch Necessary for Substrate Recognition: Crystal Structures of Lysine Epsilon-Aminotransferase (Rv3290C) from Mycobacterium Tuberculosis H37Rv.
J.Mol.Biol., 362, 2006
|
|
2KGY
 
 | | Solution structure of Rv0603 protein from Mycobacterium tuberculosis H37Rv | | Descriptor: | POSSIBLE EXPORTED PROTEIN | | Authors: | Tripathi, S, Pulavarti, S.V.S.R.K, Pathak, P.P, Meher, A.K, Jain, A, Arora, A. | | Deposit date: | 2009-03-23 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Rv0603 protein from Mycobacterium tuberculosis H37Rv
To be Published
|
|
2LRA
 
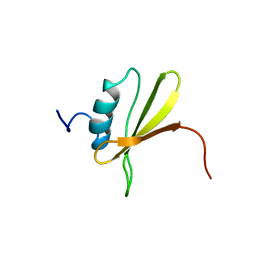 | | NMR Structure of Signal Sequence Deleted (SSD) Rv0603 Protein from Mycobacterium tuberculosis without N-terminal His-tag | | Descriptor: | POSSIBLE EXPORTED PROTEIN | | Authors: | Tripathi, S, Pulavarti, S, Yadav, R, Jain, A, Pathak, P, Meher, A, Arora, A. | | Deposit date: | 2012-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Signal Sequence Deleted (SSD) Rv0603 Protein from Mycobacterium tuberculosis without N-terminal His-tag
To be Published
|
|
2JJF
 
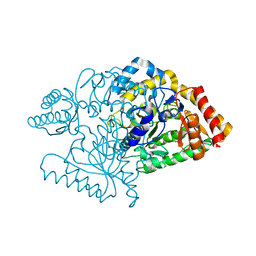 | | N328A mutant of M. tuberculosis Rv3290c | | Descriptor: | L-LYSINE EPSILON AMINOTRANSFERASE | | Authors: | tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-04-04 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational Analysis of Mycobacterium Tuberculosis Lysine Epsilon-Aminotransferase and Inhibitor Co-Crystal Structures, Reveals Distinct Binding Modes.
Biochem.Biophys.Res.Commun., 463, 2015
|
|
2JJH
 
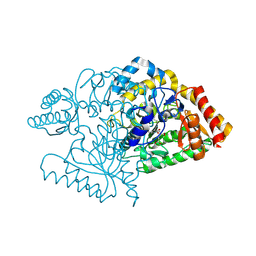 | | E243 mutant of M. tuberculosis Rv3290C | | Descriptor: | 2-OXOGLUTARIC ACID, L-LYSINE EPSILON AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-04-04 | | Release date: | 2009-06-30 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutational Analysis of Mycobacterium Tuberculosis Lysine Epsilon-Aminotransferase and Inhibitor Co-Crystal Structures, Reveals Distinct Binding Modes.
Biochem.Biophys.Res.Commun., 463, 2015
|
|
2JJE
 
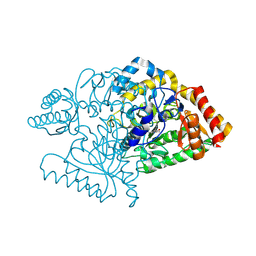 | |
2JJG
 
 | |
5ZXE
 
 | | Structure of a consensus sequence derived from the FGF family | | Descriptor: | CHLORIDE ION, Consensus sequence based basic form of fibroblast growth factor, GLYCEROL, ... | | Authors: | Tripathi, S.K, Mandalaparthy, V, Ramaswamy, S, Gosavi, S. | | Deposit date: | 2018-05-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a consensus sequence derived from the FGF family
To be published
|
|
3DZP
 
 | |
3E0A
 
 | |
3DZN
 
 | |
3DZR
 
 | |
5VZ5
 
 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2017-05-26 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5901 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
6X61
 
 | |
3DE1
 
 | |
3DDZ
 
 | |
3DE0
 
 | |
3D9Q
 
 | |
3DE2
 
 | |
3DE6
 
 | |
5ALA
 
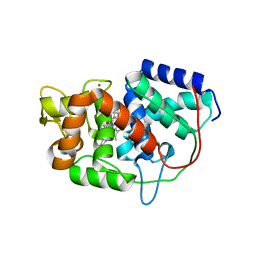 | | Structure of Leishmania major peroxidase D211R mutant (low res) | | Descriptor: | ASCORBATE PEROXIDASE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Chreifi, G, Hollingsworth, S.A, Li, H, Tripathi, S, Arce, A.P, Magana-Garcia, H.I, Poulos, T.L. | | Deposit date: | 2015-03-07 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Enzymatic Mechanism of Leishmania major Peroxidase and the Critical Role of Specific Ionic Interactions.
Biochemistry, 54, 2015
|
|
7SMD
 
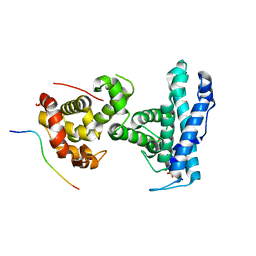 | | p107 pocket domain complexed with EID1 peptide | | Descriptor: | EP300-interacting inhibitor of differentiation 1, Retinoblastoma-like protein 1, SULFATE ION | | Authors: | Putta, S, Fernandez, S.M, Tripathi, S.M, Muller, G.A, Rubin, S.M. | | Deposit date: | 2021-10-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Structure, 30, 2022
|
|
5AL9
 
 | | Structure of Leishmania major peroxidase D211R mutant (high res) | | Descriptor: | ASCORBATE PEROXIDASE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Chreifi, G, Hollingsworth, S.A, Li, H, Tripathi, S, Arce, A.P, Magana-Garcia, H.I, Poulos, T.L. | | Deposit date: | 2015-03-07 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Enzymatic Mechanism of Leishmania major Peroxidase and the Critical Role of Specific Ionic Interactions.
Biochemistry, 54, 2015
|
|
