1MMY
 
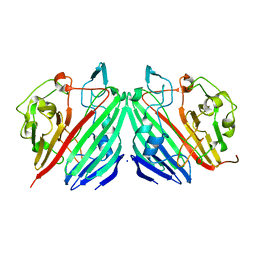 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-quinovose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, alpha-D-quinovopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1C30
 
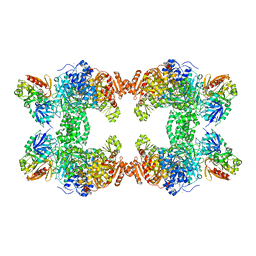 | | CRYSTAL STRUCTURE OF CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT MUTATION C269S | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-07-24 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
1CS0
 
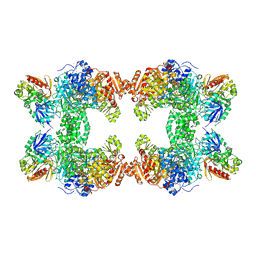 | | Crystal structure of carbamoyl phosphate synthetase complexed at CYS269 in the small subunit with the tetrahedral mimic l-glutamate gamma-semialdehyde | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-08-16 | | Release date: | 1999-12-10 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
1MMU
 
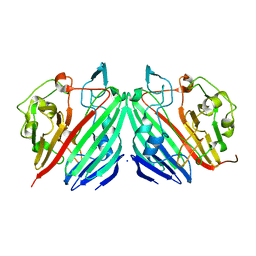 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-glucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, beta-D-glucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1KJQ
 
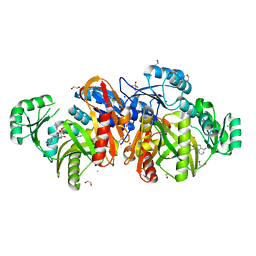 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-ADP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-05 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KJJ
 
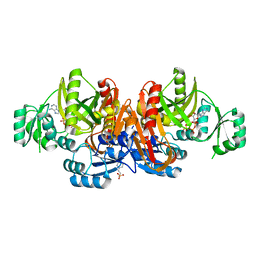 | | Crystal structure of glycniamide ribonucleotide transformylase in complex with Mg-ATP-gamma-S | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1MMZ
 
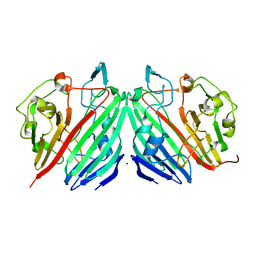 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with L-arabinose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, beta-L-arabinopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, R.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1MMX
 
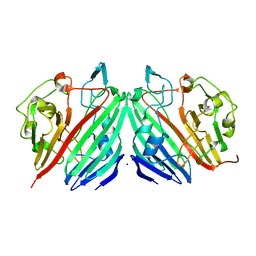 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-fucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, alpha-L-fucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1BXR
 
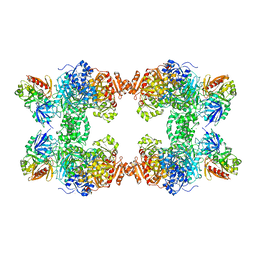 | | STRUCTURE OF CARBAMOYL PHOSPHATE SYNTHETASE COMPLEXED WITH THE ATP ANALOG AMPPNP | | Descriptor: | CARBAMOYL-PHOSPHATE SYNTHASE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Wesenberg, G, Raushel, F.M, Holden, H.M. | | Deposit date: | 1998-10-08 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl phosphate synthetase: closure of the B-domain as a result of nucleotide binding.
Biochemistry, 38, 1999
|
|
1KJI
 
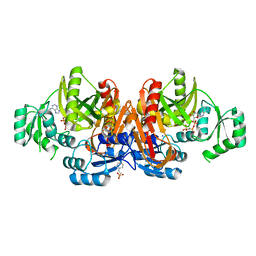 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-AMPPCP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KJ9
 
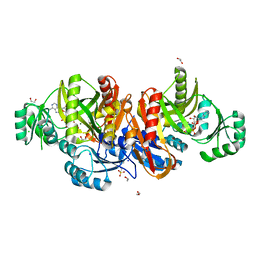 | | Crystal structure of purt-encoded glycinamide ribonucleotide transformylase complexed with Mg-ATP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KJ8
 
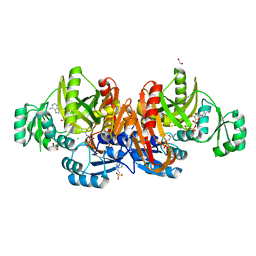 | | Crystal Structure of PurT-Encoded Glycinamide Ribonucleotide Transformylase in Complex with Mg-ATP and GAR | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1DV1
 
 | | STRUCTURE OF BIOTIN CARBOXYLASE (APO) | | Descriptor: | BIOTIN CARBOXYLASE, PHOSPHATE ION | | Authors: | Thoden, J.B, Blanchard, C.Z, Holden, H.M, Waldrop, G.L. | | Deposit date: | 2000-01-19 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Movement of the biotin carboxylase B-domain as a result of ATP binding.
J.Biol.Chem., 275, 2000
|
|
1DV2
 
 | | The structure of biotin carboxylase, mutant E288K, complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BIOTIN CARBOXYLASE | | Authors: | Thoden, J.B, Blanchard, C.Z, Holden, H.M, Waldrop, G.L. | | Deposit date: | 2000-01-19 | | Release date: | 2000-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Movement of the biotin carboxylase B-domain as a result of ATP binding.
J.Biol.Chem., 275, 2000
|
|
1M6V
 
 | | Crystal Structure of the G359F (small subunit) Point Mutant of Carbamoyl Phosphate Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-07-17 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl-phosphate synthetase. Creation of an escape route for ammonia
J.Biol.Chem., 277, 2002
|
|
1C3O
 
 | | CRYSTAL STRUCTURE OF THE CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT MUTANT C269S WITH BOUND GLUTAMINE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-07-28 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
1KVU
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1ONW
 
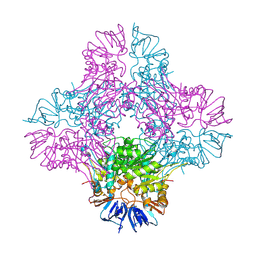 | | Crystal structure of Isoaspartyl Dipeptidase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isoaspartyl dipeptidase, ... | | Authors: | Thoden, J.B, Marti-Arbona, R, Raushel, F.M, Holden, H.M. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High Resolution X-ray Structure of
Isoaspartyl Dipeptidase from
Escherichia coli
Biochemistry, 42, 2003
|
|
1ONX
 
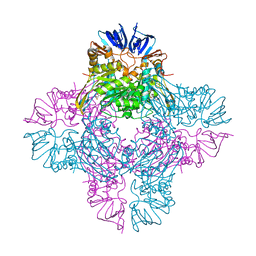 | | Crystal structure of isoaspartyl dipeptidase from escherichia coli complexed with aspartate | | Descriptor: | ASPARTIC ACID, Isoaspartyl dipeptidase, ZINC ION | | Authors: | Thoden, J.B, Marti-Arbona, R, Raushel, F.M, Holden, H.M. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution X-ray Structure of Isoaspartyl
Dipeptidase from Escherichia coli
Biochemistry, 42, 2003
|
|
1LO9
 
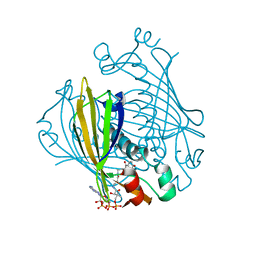 | | X-ray crystal structure of 4-hydroxybenzoyl CoA thioesterase mutant D17N complexed with 4-hydroxybenzoyl CoA | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-hydroxybenzoyl-CoA Thioesterase | | Authors: | Thoden, J.B, Holden, H.M, Zhuang, Z, Dunaway-Mariano, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase.
J.Biol.Chem., 277, 2002
|
|
1LO7
 
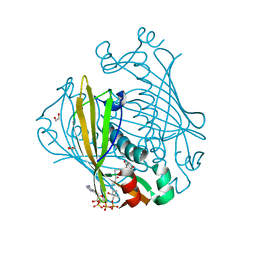 | | X-ray structure of 4-Hydroxybenzoyl CoA Thioesterase complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA Thioesterase | | Authors: | Thoden, J.B, Holden, H.M, Zhuang, Z, Dunaway-Mariano, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase.
J.Biol.Chem., 277, 2002
|
|
1LO8
 
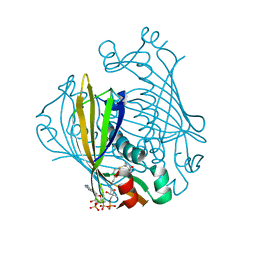 | | X-ray crystal structure of 4-hydroxybenzoyl CoA thioesterase complexed with 4-hydroxybenzyl CoA | | Descriptor: | 4-HYDROXYBENZYL COENZYME A, 4-hydroxybenzoyl-CoA Thioesterase | | Authors: | Thoden, J.B, Holden, H.M, Zhuang, Z, Dunaway-Mariano, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase.
J.Biol.Chem., 277, 2002
|
|
1A9Z
 
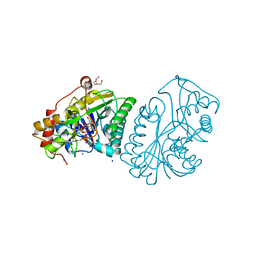 | |
1A9Y
 
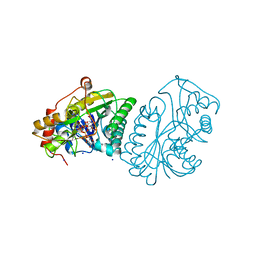 | |
1BXK
 
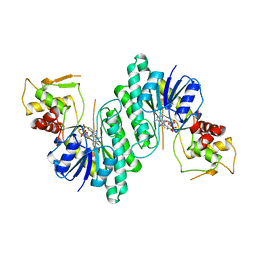 | | DTDP-GLUCOSE 4,6-DEHYDRATASE FROM E. COLI | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (DTDP-GLUCOSE 4,6-DEHYDRATASE) | | Authors: | Thoden, J.B, Hegeman, A.D, Frey, P.A, Holden, H.M. | | Deposit date: | 1998-10-05 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Structure of Dtdp-Glucose 4,6-Dehydratase from E. Coli
Protein Sci.
|
|
