2BM9
 
 | | cmcI-N160 in complex with SAM | | Descriptor: | CEPHALOSPORIN HYDROXYLASE CMCI, S-ADENOSYLMETHIONINE | | Authors: | Oster, L.M, Lester, D.R, Terwisscha Van Scheltinga, A, Svenda, M, Genereux, C, Andersson, I. | | Deposit date: | 2005-03-10 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Insights Into Cephamycin Biosynthesis: The Crystal Structure of Cmci from Streptomyces Clavuligerus
J.Mol.Biol., 358, 2006
|
|
2BR4
 
 | | cmcI-D160 Mg-SAM | | Descriptor: | CEPHALOSPORIN HYDROXYLASE CMCI, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Oster, L.M, Lester, D.R, Terwisscha van Scheltinga, A, Svenda, M, Genereux, C, Andersson, I. | | Deposit date: | 2005-05-01 | | Release date: | 2006-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights Into Cephamycin Biosynthesis: The Crystal Structure of Cmci from Streptomyces Clavuligerus.
J.Mol.Biol., 358, 2006
|
|
2BM8
 
 | | CmcI-N160 apo-structure | | Descriptor: | CEPHALOSPORIN HYDROXYLASE CMCI | | Authors: | Oster, L.M, Lester, D.R, Terwisscha van Scheltinga, A, Svenda, M, Genereux, C, Andersson, I. | | Deposit date: | 2005-03-10 | | Release date: | 2006-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Cephamycin Biosynthesis: The Crystal Structure of Cmci from Streptomyces Clavuligerus
J.Mol.Biol., 358, 2006
|
|
2BR5
 
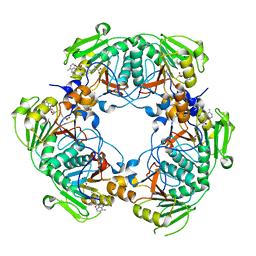 | | cmcI-N160 SAH | | Descriptor: | CEPHALOSPORIN HYDROXYLASE CMCI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Oster, L.M, Lester, D.R, Terwisscha van Scheltinga, A, svenda, M, Genereux, C, Andersson, I. | | Deposit date: | 2005-05-01 | | Release date: | 2006-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Insights Into Cephamycin Biosynthesis: The Crystal Structure of Cmci from Streptomyces Clavuligerus.
J.Mol.Biol., 358, 2006
|
|
2BHW
 
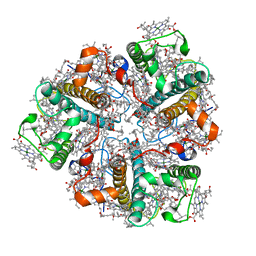 | | PEA LIGHT-HARVESTING COMPLEX II AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6'S,9R,9'R,13R,13'S)-4',5'-DIDEHYDRO-5',6',7',8',9,9',10,10',11,11',12,12',13,13',14,14',15,15'-OCTADECAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Standfuss, J, Terwisscha van Scheltinga, A.C, Lamborghini, M, Kuehlbrandt, W. | | Deposit date: | 2005-01-19 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms of Photoprotection and Nonphotochemical Quenching in Pea Light-Harvesting Complex at 2.5 A Resolution.
Embo J., 24, 2005
|
|
2VPN
 
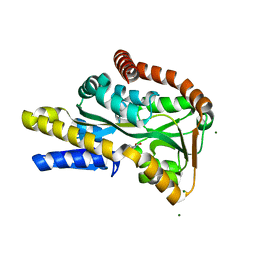 | | High-resolution structure of the periplasmic ectoine-binding protein from TeaABC TRAP-transporter of Halomonas elongata | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, MAGNESIUM ION, PERIPLASMIC SUBSTRATE BINDING PROTEIN | | Authors: | Kuhlmann, S.I, Terwisscha van Scheltinga, A.C, Bienert, R, Kunte, H.J, Ziegler, C. | | Deposit date: | 2008-03-03 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 A Structure of the Ectoine Binding Protein Teaa of the Osmoregulated Trap-Transporter Teaabc from Halomonas Elongata.
Biochemistry, 47, 2008
|
|
2WSW
 
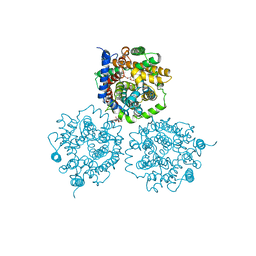 | | Crystal Structure of Carnitine Transporter from Proteus mirabilis | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, BCCT FAMILY BETAINE/CARNITINE/CHOLINE TRANSPORTER, GLYCEROL, ... | | Authors: | Schulze, S, Terwisscha van Scheltinga, A.C, Kuehlbrandt, W. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural Basis of Na(+)-Independent and Cooperative Substrate/Product Antiport in Cait.
Nature, 467, 2010
|
|
2WIT
 
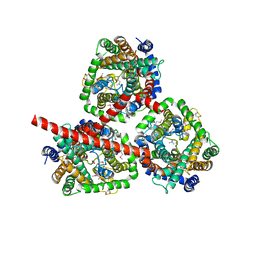 | | CRYSTAL STRUCTURE OF THE SODIUM-COUPLED GLYCINE BETAINE SYMPORTER BETP FROM CORYNEBACTERIUM GLUTAMICUM WITH BOUND SUBSTRATE | | Descriptor: | GLYCINE BETAINE TRANSPORTER BETP, TRIMETHYL GLYCINE | | Authors: | Ressl, S, Terwisscha Van Scheltinga, A.C, Vonrhein, C, Ott, V, Ziegler, C. | | Deposit date: | 2009-05-17 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Molecular Basis of Transport and Regulation in the Na(+)/Betaine Symporter Betp.
Nature, 458, 2009
|
|
2WSX
 
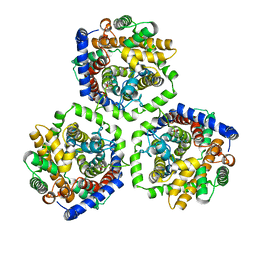 | | Crystal Structure of Carnitine Transporter from Escherichia coli | | Descriptor: | 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, L-CARNITINE/GAMMA-BUTYROBETAINE ANTIPORTER | | Authors: | Schulze, S, Terwisscha van Scheltinga, A.C, Kuehlbrandt, W. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Na(+)-Independent and Cooperative Substrate/Product Antiport in Cait.
Nature, 467, 2010
|
|
2VPO
 
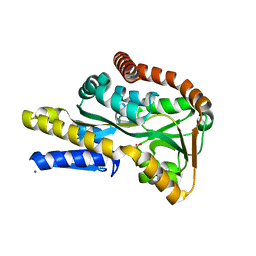 | | High resolution structure of the periplasmic binding protein TeaA from TeaABC TRAP transporter of Halomonas elongata in complex with hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, MAGNESIUM ION, PERIPLASMIC SUBSTRATE BINDING PROTEIN | | Authors: | Kuhlmann, S.I, Terwisscha van Scheltinga, A.C, Bienert, R, Kunte, H.J, Ziegler, C. | | Deposit date: | 2008-03-03 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.55 A Structure of the Ectoine Binding Protein Teaa of the Osmoregulated Trap-Transporter Teaabc from Halomonas Elongata.
Biochemistry, 47, 2008
|
|
