5XNR
 
 | | Truncated AlyQ with CBM32 and alginate lyase domains | | Descriptor: | ACETATE ION, AlyQ, CALCIUM ION, ... | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and Structural Studies of a Multidomain Alginate Lyase from Persicobacter sp. CCB-QB2.
Sci Rep, 7, 2017
|
|
6AII
 
 | | Catalytic domain of PdAgaC | | Descriptor: | Beta-agarase, CALCIUM ION | | Authors: | Teh, A.H, Fazli, N.H. | | Deposit date: | 2018-08-23 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of a neoagarobiose-producing GH16 family beta-agarase from Persicobacter sp. CCB-QB2.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
2FR6
 
 | | Crystal Structure of Mouse Cytidine Deaminase Complexed with Cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, AMMONIA, Cytidine deaminase, ... | | Authors: | Teh, A.H. | | Deposit date: | 2006-01-19 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The 1.48 A Resolution Crystal Structure of the Homotetrameric Cytidine Deaminase from Mouse
Biochemistry, 45, 2006
|
|
2FR5
 
 | |
7D2A
 
 | | CBM32 of AlyQ in complex with 4,5-unsaturated mannuronic acid | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, AlyQ, CALCIUM ION, ... | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for binding uronic acids by family 32 carbohydrate-binding modules.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7D29
 
 | | CBM32 of AlyQ | | Descriptor: | AlyQ, CALCIUM ION | | Authors: | Teh, A.H, Sim, P.F. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for binding uronic acids by family 32 carbohydrate-binding modules.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
3S1I
 
 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S1J
 
 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
1ZAB
 
 | | Crystal Structure of Mouse Cytidine Deaminase Complexed with 3-Deazauridine | | Descriptor: | 1-((2R,3R,4S,5R)-TETRAHYDRO-3,4-DIHYDROXY-5-(HYDROXYMETHYL)FURAN-2-YL)PYRIDINE-2,4(1H,3H)-DIONE, Cytidine deaminase, SULFATE ION, ... | | Authors: | Teh, A.H. | | Deposit date: | 2005-04-06 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The 1.48 A Resolution Crystal Structure of the Homotetrameric Cytidine Deaminase from Mouse
Biochemistry, 45, 2006
|
|
3W41
 
 | | Crystal structure of RsbX in complex with magnesium in space group P21 | | Descriptor: | MAGNESIUM ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W42
 
 | | Crystal structure of RsbX in complex with manganese in space group P1 | | Descriptor: | MANGANESE (II) ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W44
 
 | | Crystal structure of RsbX, selenomethionine derivative | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W40
 
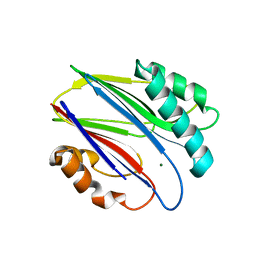 | | Crystal structure of RsbX in complex with magnesium in space group P1 | | Descriptor: | MAGNESIUM ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WFW
 
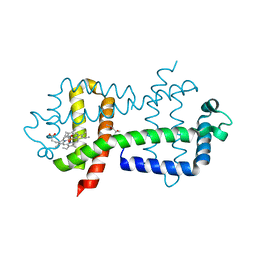 | | Crystal Structure of the Closed Form of the HGbRL's Globin Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hemoglobin-like flavoprotein fused to Roadblock/LC7 domain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Teh, A.H. | | Deposit date: | 2013-07-24 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Open and Lys-His Hexacoordinated Closed Structures of a Globin with Swapped Proximal and Distal Sites.
Sci Rep, 5, 2015
|
|
3WFX
 
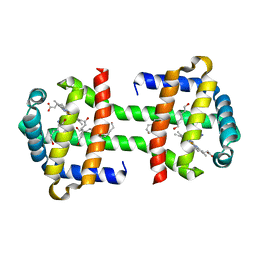 | | Crystal Structure of the Imidazole-Bound Form of the HGbRL's Globin Domain | | Descriptor: | HEXANE-1,6-DIOL, Hemoglobin-like flavoprotein fused to Roadblock/LC7 domain, IMIDAZOLE, ... | | Authors: | Teh, A.H. | | Deposit date: | 2013-07-24 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Open and Lys-His Hexacoordinated Closed Structures of a Globin with Swapped Proximal and Distal Sites.
Sci Rep, 5, 2015
|
|
3W43
 
 | | Crystal structure of RsbX in complex with manganese in space group P21 | | Descriptor: | MANGANESE (II) ION, Phosphoserine phosphatase RsbX | | Authors: | Teh, A.H, Makino, M, Baba, S, Shimizu, N, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2013-01-04 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2Z3J
 
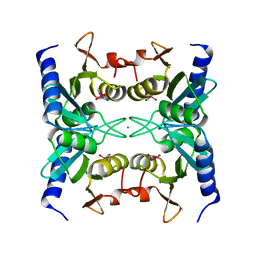 | | Crystal structure of blasticidin S deaminase (BSD) R90K mutant | | Descriptor: | Blasticidin-S deaminase, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Teh, A.H, Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
3CE1
 
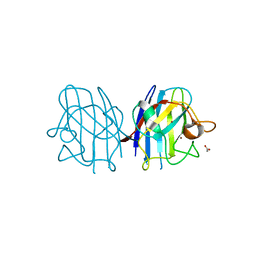 | | Crystal Structure of the Cu/Zn Superoxide Dismutase from Cryptococcus liquefaciens Strain N6 | | Descriptor: | ACETATE ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Teh, A.H, Kanamasa, S, Kajiwara, S, Kumasaka, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Cu/Zn superoxide dismutase from the heavy-metal-tolerant yeast Cryptococcus liquefaciens strain N6.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
5ZRT
 
 | | Crystal structure of human C1ORF123 protein | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rahaman, S.N.A, Yusop, J.M, Mohamed-Hussein, Z.A, Wan Mohd, A, Ho, K.L, Teh, A.H, Waterman, J, Ng, C.L. | | Deposit date: | 2018-04-25 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of human C1ORF123.
Peerj, 6, 2018
|
|
5YNX
 
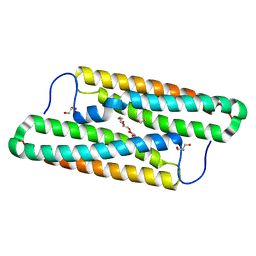 | | Structure of house dust mite allergen Der f 21 in PEG400 | | Descriptor: | Allergen Der f 21, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I, Beis, K, Say, Y.H. | | Deposit date: | 2017-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
5YNY
 
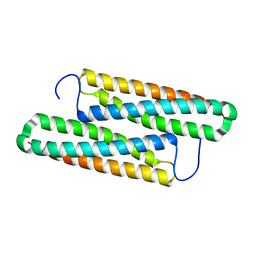 | | Structure of house dust mite allergen Der F 21 in PEG2KMME | | Descriptor: | Allergen Der f 21 | | Authors: | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I. | | Deposit date: | 2017-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
4E2O
 
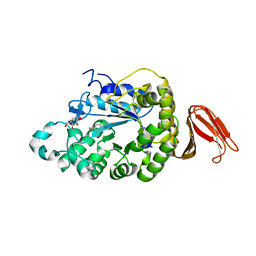 | | Crystal structure of alpha-amylase from Geobacillus thermoleovorans, GTA, complexed with acarbose | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, CALCIUM ION, ... | | Authors: | Mok, S.C, Teh, A.H, Saito, J.A, Najimudin, N, Alam, M. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of a compact alpha-amylase from Geobacillus thermoleovorans.
Enzyme.Microb.Technol., 53, 2013
|
|
5A2A
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ACETATE ION, APO FORM OF ANOXYBACILLUS ALPHA-AMYLASES, CALCIUM ION | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-16 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5A2B
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ANOXYBACILLUS ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5A2C
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
