1VDI
 
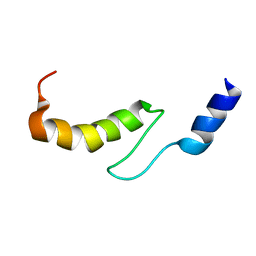 | | Solution structure of actin-binding domain of troponin in Ca2+-free state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
1VDJ
 
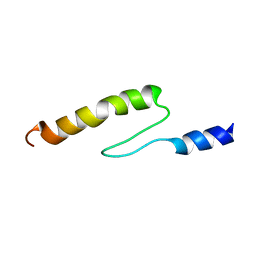 | | Solution structure of actin-binding domain of troponin in Ca2+-bound state | | Descriptor: | Troponin I, fast skeletal muscle | | Authors: | Murakami, K, Yumoto, F, Ohki, S, Yasunaga, T, Tanokura, M, Wakabayashi, T. | | Deposit date: | 2004-03-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ca(2+)-regulated Muscle Relaxation at Interaction Sites of Troponin with Actin and Tropomyosin
J.Mol.Biol., 352, 2005
|
|
4YOV
 
 | |
4YOX
 
 | |
4H7E
 
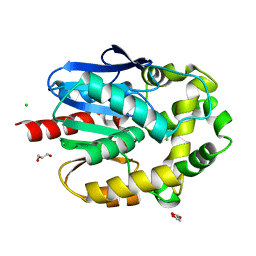 | | Crystal structure of haloalkane dehalogenase LinB V112A mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7F
 
 | | Crystal structure of haloalkane dehalogenase LinB V134I mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
2EGD
 
 | | Crystal structure of human S100A13 in the Ca2+-bound state | | Descriptor: | CALCIUM ION, Protein S100-A13 | | Authors: | Imai, F.L, Nagata, K, Yonezawa, N, Nakano, M, Tanokura, M. | | Deposit date: | 2007-02-28 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human S100A13 in the Ca2+-bound state
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4H8N
 
 | | Crystal structure of conjugated polyketone reductase C2 from candida parapsilosis complexed with NADPH | | Descriptor: | Conjugated polyketone reductase C2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
1Y49
 
 | |
4H7D
 
 | | Crystal structure of haloalkane dehalogenase LinB T81A mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
5B0H
 
 | | CRYSTAL STRUCTURE OF HUMAN LEUKOCYTE CELL-DERIVED CHEMOTAXIN 2 | | Descriptor: | Leukocyte cell-derived chemotaxin-2, SULFATE ION, ZINC ION | | Authors: | Zheng, H, Miyakawa, T, Sawano, Y, Tanokura, M. | | Deposit date: | 2015-10-29 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Human Leukocyte Cell-derived Chemotaxin 2 (LECT2) Reveals a Mechanistic Basis of Functional Evolution in a Mammalian Protein with an M23 Metalloendopeptidase Fold
J.Biol.Chem., 291, 2016
|
|
4H77
 
 | | Crystal structure of haloalkane dehalogenase LinB from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, F.L, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7J
 
 | | Crystal structure of haloalkane dehalogenase LinB H247A mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7K
 
 | | Crystal structure of haloalkane dehalogenase LinB I253M mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
1ZR7
 
 | |
4H7H
 
 | | Crystal structure of haloalkane dehalogenase LinB T135A mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7I
 
 | | Crystal structure of haloalkane dehalogenase LinB L138I mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
7CFA
 
 | |
7CO1
 
 | | Crystal structure of SMAD2 in complex with wild-type CBP | | Descriptor: | CREB-binding protein, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Wada, H, Ito, T, Tanokura, M. | | Deposit date: | 2020-08-03 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
1V5Y
 
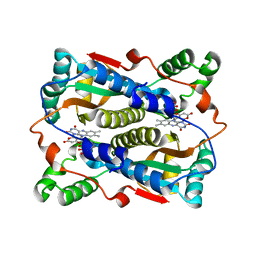 | | Binding of coumarins to NAD(P)H:FMN oxidoreductase | | Descriptor: | 4-HYDROXY-2H-CHROMEN-2-ONE, FLAVIN MONONUCLEOTIDE, Major NAD(P)H-flavin oxidoreductase | | Authors: | Kobori, T, Koike, H, Sasaki, H, Zenno, S, Saigo, K, Tanokura, M. | | Deposit date: | 2003-11-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of coumarins to NAD(P)H:FMN oxidoreductase
To be Published
|
|
1V3Z
 
 | |
1V46
 
 | |
1V5Z
 
 | | Binding of coumarins to NAD(P)H:FMN oxidoreductase | | Descriptor: | (2E)-3-(2-HYDROXYPHENYL)ACRYLIC ACID, FLAVIN MONONUCLEOTIDE, Major NAD(P)H-flavin oxidoreductase | | Authors: | Kobori, T, Koike, H, Sasaki, H, Zenno, S, Saigo, K, Tanokura, M. | | Deposit date: | 2003-11-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of coumarins to NAD(P)H:FMN oxidoreductase
To be Published
|
|
4HE7
 
 | | Crystal Structure of Brazzein | | Descriptor: | Defensin-like protein, SODIUM ION | | Authors: | Nagata, K, Hongo, N, Kameda, Y, Yamamura, A, Sasaki, H, Lee, W.C, Ishikawa, K, Suzuki, E, Tanokura, M. | | Deposit date: | 2012-10-03 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5XOC
 
 | | Crystal structure of human Smad3-FoxH1 complex | | Descriptor: | Mothers against decapentaplegic homolog 3, Thioredoxin 1,Forkhead box protein H1 | | Authors: | Miyazono, K, Ito, T, Tanokura, M. | | Deposit date: | 2017-05-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors
Sci Signal, 11, 2018
|
|
