7W2J
 
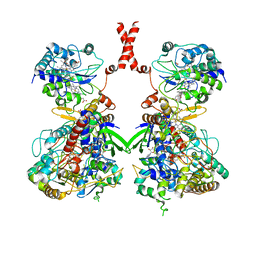 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
7WSQ
 
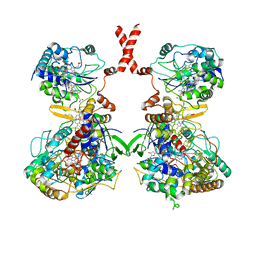 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
7BZK
 
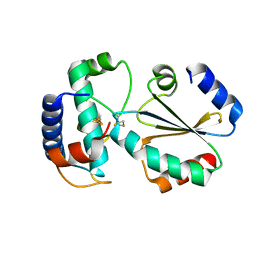 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin y1 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, IRON/SULFUR CLUSTER, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5935 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7BW2
 
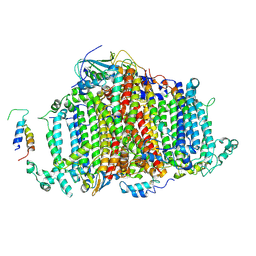 | | Crystal Structure of Cyanobacterial PSI Monomer from T.elongatus at 6.5 A Resolution | | Descriptor: | Photosystem I 4.8K protein, Photosystem I P700 chlorophyll a apoprotein A1, Photosystem I P700 chlorophyll a apoprotein A2, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Eithar, E.M, Mian, Y. | | Deposit date: | 2020-04-13 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
7C65
 
 | | Crystal structure of thioredoxin m1 | | Descriptor: | SODIUM ION, Thioredoxin M1, chloroplastic | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7C2B
 
 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin f2 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, Ferredoxin-thioredoxin reductase variable chain, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7949 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7C3F
 
 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin m2 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, Ferredoxin-thioredoxin reductase variable chain, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3986 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7CKA
 
 | |
6J13
 
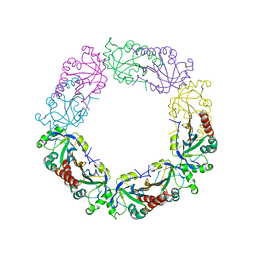 | | Redox protein from Chlamydomonas reinhardtii | | Descriptor: | 2-cys peroxiredoxin | | Authors: | Charoenwattansatien, R, Zinzius, K, Tanaka, H, Hippler, M, Kurisu, G. | | Deposit date: | 2018-12-27 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green algaChlamydomonas reinhardtii.
J.Biol.Chem., 295, 2020
|
|
6L4P
 
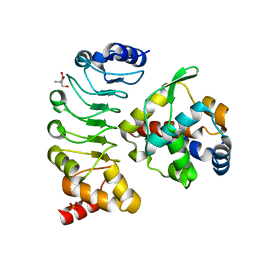 | | Crystal structure of the complex between the axonemal outer-arm dynein light chain-1 and microtubule binding domain of gamma heavy chain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dynein light chain 1, axonemal, ... | | Authors: | Toda, A, Nishikawa, Y, Tanaka, H, Yagi, T, Kurisu, G. | | Deposit date: | 2019-10-19 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The complex of outer-arm dynein light chain-1 and the microtubule-binding domain of the gamma heavy chain shows how axonemal dynein tunes ciliary beating.
J.Biol.Chem., 295, 2020
|
|
3X2L
 
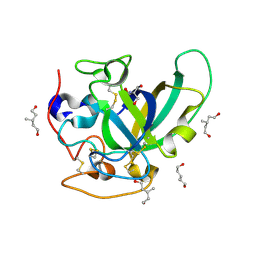 | | X-ray structure of PcCel45A apo form at 95K. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
6LU1
 
 | | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Gerle, C, Kawamoto, A, Kato, T, Namba, K, Nowaczyk, M.M, Rogner, M, Misumi, Y, Frank, A, Eithar, E.M. | | Deposit date: | 2020-01-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
5Y9Z
 
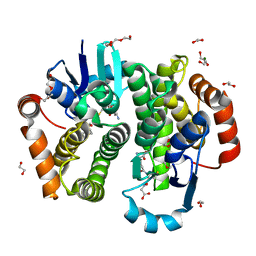 | | Crystal structure of rat hematopoietic prostaglandin D synthase | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, GLUTATHIONE, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Tanaka, H, Aritake, K, Urade, Y. | | Deposit date: | 2017-08-29 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal structure of rat hematopoietic prostaglandin D synthase
To Be Published
|
|
5YXM
 
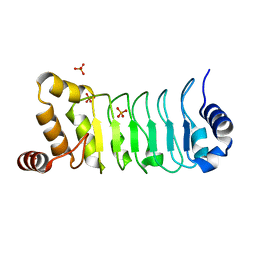 | | Crystal structure of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Dynein light chain 1, axonemal, PHOSPHATE ION | | Authors: | Toda, A, Tanaka, H, Nishikawa, Y, Yagi, T, Kurisu, G. | | Deposit date: | 2017-12-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Structural atlas of dynein motors at atomic resolution.
Biophys Rev, 10, 2018
|
|
6AA2
 
 | | X-ray structure of ReQy1 (oxidized form) | | Descriptor: | Green fluorescent protein | | Authors: | Sugiura, K, Yasuda, A, Tabushi, N, Tanaka, H, Kurisu, G, Hisabori, T. | | Deposit date: | 2018-07-17 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multicolor redox sensor proteins can visualize redox changes in various compartments of the living cell.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
1UKJ
 
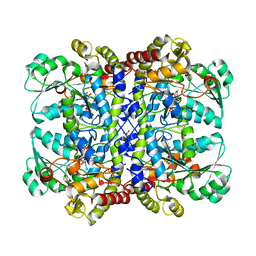 | | Detailed structure of L-Methionine-Lyase from Pseudomonas putida | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Misaki, S, Takimoto, A, Takakura, T, Yoshioka, T, Yamashita, M, Tamura, T, Tanaka, H, Inagaki, K. | | Deposit date: | 2003-08-24 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Detailed structure of L-Methionine -Lyase from Pseudomonas putida
To be Published
|
|
2E0Z
 
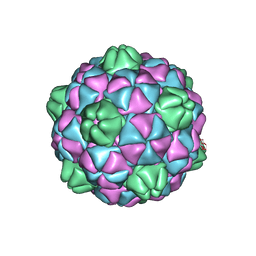 | | Crystal structure of virus-like particle from Pyrococcus furiosus | | Descriptor: | Virus-like particle | | Authors: | Akita, F, Chong, K.T, Tanaka, H, Yamashita, E, Miyazaki, N, Nakaishi, Y, Namba, K, Ono, Y, Suzuki, M, Tsukihara, T, Nakagawa, A. | | Deposit date: | 2006-10-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses
J.Mol.Biol., 368, 2007
|
|
2O7C
 
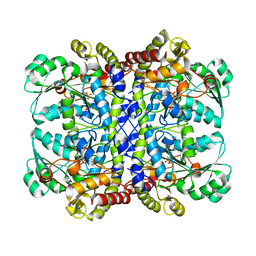 | | Crystal structure of L-methionine-lyase from Pseudomonas | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Misaki, S, Takimoto, A, Takakura, T, Yoshioka, T, Yamashita, M, Tamura, T, Tanaka, H, Inagaki, K. | | Deposit date: | 2006-12-10 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the antitumour enzyme L-methionine gamma-lyase from Pseudomonas putida at 1.8 A resolution
J.Biochem.(Tokyo), 141, 2007
|
|
3ZGF
 
 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with in complex with NPE caged UDP-Gal (P2(1)2(1)2(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3ZGG
 
 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with NPE caged UDP-Gal (C222(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, GLYCEROL, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
