1TUW
 
 | | Structural and Functional Analysis of Tetracenomycin F2 Cyclase from Streptomyces glaucescens: A Type-II Polyketide Cyclase | | 分子名称: | SULFATE ION, Tetracenomycin polyketide synthesis protein tcmI | | 著者 | Thompson, T.B, Katayama, K, Watanabe, K, Hutchinson, C.R, Rayment, I. | | 登録日 | 2004-06-25 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional analysis of tetracenomycin F2 cyclase from Streptomyces glaucescens. A type II polyketide cyclase.
J.Biol.Chem., 279, 2004
|
|
1UB8
 
 | | Crystal structure of d(GCGAAGC), bending duplex with a bulge-in residue | | 分子名称: | 5'-D(*GP*CP*GP*AP*AP*GP*C)-3', COBALT HEXAMMINE(III) | | 著者 | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | 登録日 | 2003-03-31 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IP1
 
 | | G37A HUMAN LYSOZYME | | 分子名称: | LYSOZYME C, SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2001-04-20 | | 公開日 | 2001-11-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP4
 
 | | G72A HUMAN LYSOZYME | | 分子名称: | LYSOZYME C, SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2001-04-20 | | 公開日 | 2001-11-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP3
 
 | | G68A HUMAN LYSOZYME | | 分子名称: | LYSOZYME C, SODIUM ION, SULFATE ION | | 著者 | Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2001-04-20 | | 公開日 | 2001-11-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP2
 
 | | G48A HUMAN LYSOZYME | | 分子名称: | LYSOZYME C, SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2001-04-20 | | 公開日 | 2001-11-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IX0
 
 | |
7YRK
 
 | | Crystal structure of the hen egg lysozyme-2-oxidobenzylidene-threoninato-copper (II) complex | | 分子名称: | (6~{S})-6-[(1~{R})-1-oxidanylethyl]-2,4-dioxa-7$l^{4}-aza-3$l^{3}-cupratricyclo[7.4.0.0^{3,7}]trideca-1(13),7,9,11-tetraen-5-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Unno, M, Furuya, T, Kitanishi, K, Akitsu, T. | | 登録日 | 2022-08-10 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.92 Å) | | 主引用文献 | A novel hybrid protein composed of superoxide-dismutase-active Cu(II) complex and lysozyme.
Sci Rep, 13, 2023
|
|
6KAW
 
 | | Crystal structure of CghA | | 分子名称: | CghA | | 著者 | Hara, K, Hashimoto, H, Yokoyama, M, Sato, M, Watanabe, K. | | 登録日 | 2019-06-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels-Alderase
Nat Catal, 2021
|
|
3WSO
 
 | | Crystal structure of the Skp1-FBG3 complex | | 分子名称: | F-box only protein 44, S-phase kinase-associated protein 1 | | 著者 | Kumanomidou, T, Nishio, K, Takagi, K, Nakagawa, T, Suzuki, A, Yamane, T, Tokunaga, F, Iwai, K, Murakami, A, Yoshida, Y, Tanaka, K, Mizushima, T. | | 登録日 | 2014-03-18 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Structural Differences between a Glycoprotein Specific F-Box Protein Fbs1 and Its Homologous Protein FBG3
Plos One, 10, 2015
|
|
5GV1
 
 | | Crystal structure of ENZbleach xylanase wild type | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Chitnumsub, P, Jaruwat, A, Boonyapakorn, K, Noytanom, K. | | 登録日 | 2016-09-01 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-based protein engineering for thermostable and alkaliphilic enhancement of endo-beta-1,4-xylanase for applications in pulp bleaching
J. Biotechnol., 259, 2017
|
|
1INU
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1IP5
 
 | | G105A HUMAN LYSOZYME | | 分子名称: | LYSOZYME C, SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2001-04-20 | | 公開日 | 2001-11-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
1IP7
 
 | | G129A HUMAN LYSOZYME | | 分子名称: | LYSOZYME C, SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2001-04-20 | | 公開日 | 2001-11-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
6KBC
 
 | | Crystal structure of CghA with Sch210972 | | 分子名称: | (2S)-3-[(2S,4E)-4-[[(1R,2S,4aR,6S,8R,8aS)-2-[(E)-but-2-en-2-yl]-6,8-dimethyl-1,2,4a,5,6,7,8,8a-octahydronaphthalen-1-yl]-oxidanyl-methylidene]-3,5-bis(oxidanylidene)pyrrolidin-2-yl]-2-methyl-2-oxidanyl-propanoic acid, CghA | | 著者 | Hara, K, Hashimoto, H, Maeda, N, Sato, M, Watanabe, K. | | 登録日 | 2019-06-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels-Alderase
Nat Catal, 2021
|
|
2D57
 
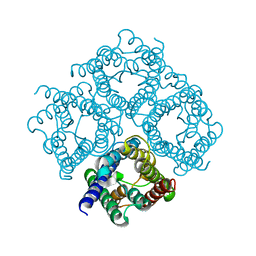 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | 分子名称: | Aquaporin-4 | | 著者 | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | 登録日 | 2005-10-29 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | 主引用文献 | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
1GE4
 
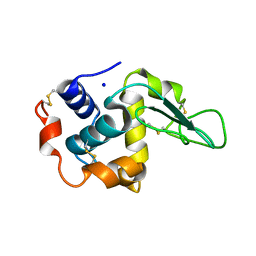 | |
1GFG
 
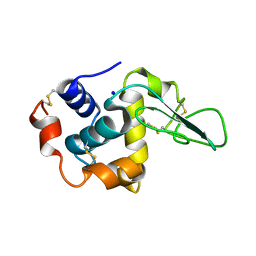 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFR
 
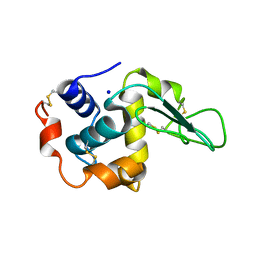 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1MH6
 
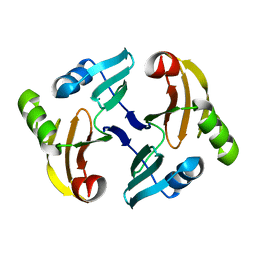 | | Solution Structure of the Transposon Tn5-encoding Bleomycin-binding Protein, BLMT | | 分子名称: | BLEOMYCIN RESISTANCE PROTEIN | | 著者 | Kumagai, T, Ohtani, K, Tsuboi, Y, Koike, T, Sugiyama, M. | | 登録日 | 2002-08-19 | | 公開日 | 2003-02-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the transposon Tn5-encoding bleomycin-binding protein complexed with an activated bleomycin analogue.
To be published
|
|
1IP6
 
 | | G127A HUMAN LYSOZYME | | 分子名称: | LYSOZYME C, SODIUM ION | | 著者 | Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2001-04-20 | | 公開日 | 2001-11-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of amino acid residues in left-handed helical conformation for the conformational stability of a protein.
Proteins, 45, 2001
|
|
2DH6
 
 | | Crystal structure of E. coli Apo-TrpB | | 分子名称: | SULFATE ION, tryptophan synthase beta subunit | | 著者 | Nishio, K, Morimoto, Y, Ogasahara, K, Yasuoka, N, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2007-04-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding
Febs J., 277, 2010
|
|
2DH5
 
 | | Crystal structure of E. coli Holo-TrpB | | 分子名称: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | 著者 | Nishio, K, Morimoto, Y, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2007-04-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding
Febs J., 277, 2010
|
|
1UE2
 
 | | Crystal structure of d(GC38GAAAGCT) | | 分子名称: | 5'-D(*GP*(C38)P*GP*AP*AP*AP*GP*CP*T)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | 著者 | Sunami, T, Kondo, J, Hirao, I, Watanaba, K, Miura, K, Takenaka, A. | | 登録日 | 2003-05-08 | | 公開日 | 2004-01-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1Z8T
 
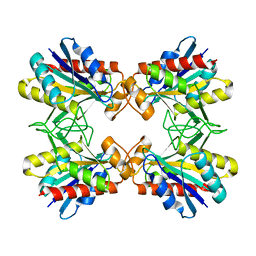 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192Q) from a Hyperthermophile, Pyrococcus furiosus | | 分子名称: | Pyrrolidone-carboxylate peptidase | | 著者 | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | 登録日 | 2005-03-31 | | 公開日 | 2006-06-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
