1IYF
 
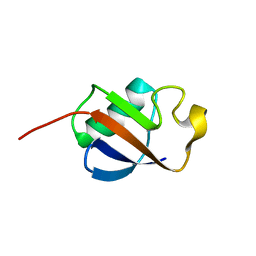 | | Solution structure of ubiquitin-like domain of human parkin | | 分子名称: | parkin | | 著者 | Sakata, E, Yamaguchi, Y, Kurimoto, E, Kikuchi, J, Yokoyama, S, Kawahara, H, Yokosawa, H, Hattori, N, Mizuno, Y, Tanaka, K, Kato, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-08-13 | | 公開日 | 2003-03-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain
EMBO REP., 4, 2003
|
|
1UMH
 
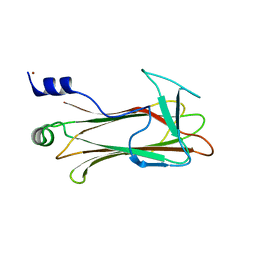 | | Structural basis of sugar-recognizing ubiquitin ligase | | 分子名称: | F-box only protein 2, NICKEL (II) ION | | 著者 | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-10-01 | | 公開日 | 2004-04-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of sugar-recognizing ubiquitin ligase
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1GEZ
 
 | |
1GF4
 
 | |
1DI4
 
 | |
1X23
 
 | | Crystal structure of ubch5c | | 分子名称: | Ubiquitin-conjugating enzyme E2 D3 | | 著者 | Nakanishi, M, Teshima, N, Mizushima, T, Murata, S, Tanaka, K, Yamane, T. | | 登録日 | 2005-04-19 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of ubch5c
To be Published
|
|
1WZV
 
 | | Crystal Structure of UbcH8 | | 分子名称: | Ubiquitin-conjugating enzyme E2 L6 | | 著者 | Mizushima, T, Suzuki, M, Teshima, N, Yamane, T, Murata, S, Tanaka, K. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of UbcH8
To be Published
|
|
5KNC
 
 | | Crystal structure of the 3 ADP-bound V1 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | 登録日 | 2016-06-28 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.015 Å) | | 主引用文献 | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
1WZW
 
 | | Crystal Structure of UbcH8 | | 分子名称: | Ubiquitin-conjugating enzyme E2 L6 | | 著者 | Mizushima, T, Suzuki, M, Teshima, N, Yamane, T, Murata, S, Tanaka, K. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of UbcH8
To be Published
|
|
5KNB
 
 | | Crystal structure of the 2 ADP-bound V1 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | 登録日 | 2016-06-28 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.251 Å) | | 主引用文献 | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KND
 
 | | Crystal structure of the Pi-bound V1 complex | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | 登録日 | 2016-06-28 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.888 Å) | | 主引用文献 | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5WTQ
 
 | | Crystal structure of human proteasome-assembling chaperone PAC4 | | 分子名称: | CHLORIDE ION, NICKEL (II) ION, Proteasome assembly chaperone 4 | | 著者 | Kurimoto, E, Satoh, T, Ito, Y, Ishihara, E, Tanaka, K, Kato, K. | | 登録日 | 2016-12-13 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human proteasome assembly chaperone PAC4 involved in proteasome formation
Protein Sci., 26, 2017
|
|
5FJJ
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | 著者 | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | 登録日 | 2015-10-09 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | 著者 | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | 登録日 | 2015-10-09 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
1GE1
 
 | |
1GB7
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-06-26 | | 公開日 | 2000-07-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GBX
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-06-26 | | 公開日 | 2000-07-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GB2
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-06-26 | | 公開日 | 2000-07-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GB9
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-06-26 | | 公開日 | 2000-07-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GF8
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GAZ
 
 | | Crystal Structure of Mutant Human Lysozyme Substituted at the Surface Positions | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-06-26 | | 公開日 | 2000-07-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of amino acid residues at turns in the conformational stability and folding of human lysozyme.
Biochemistry, 39, 2000
|
|
1GB5
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-06-26 | | 公開日 | 2000-07-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GFE
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GBO
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-06-26 | | 公開日 | 2000-07-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
1GE0
 
 | |
