7WKF
 
 | | Antimicrobial peptide-LaIT2 | | Descriptor: | Beta-KTx-like peptide LaIT2 | | Authors: | Tamura, M, Morita, H, Ohki, S. | | Deposit date: | 2022-01-09 | | Release date: | 2023-04-05 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of LaIT2, an antimicrobial and insecticidal peptide from Liocheles australasiae.
Toxicon, 214, 2022
|
|
3VOH
 
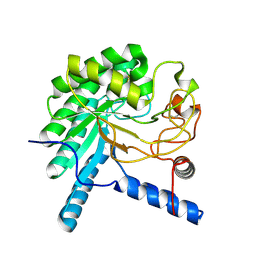 | | CcCel6A catalytic domain complexed with cellobiose | | Descriptor: | Cellobiohydrolase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOJ
 
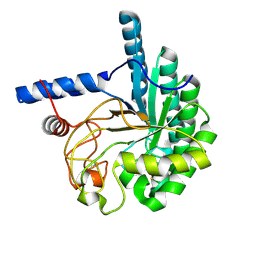 | | CcCel6A catalytic domain mutant D164A | | Descriptor: | Cellobiohydrolase | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOG
 
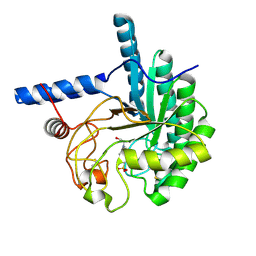 | | Catalytic domain of the cellobiohydrolase, CcCel6A, from Coprinopsis cinerea | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cellobiohydrolase | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOI
 
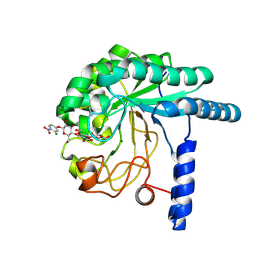 | | CcCel6A catalytic domain complexed with p-nitrophenyl beta-D-cellotrioside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
3VOF
 
 | | Cellobiohydrolase mutant, CcCel6C D102A, in the closed form | | Descriptor: | Cellobiohydrolase, beta-D-glucopyranose | | Authors: | Tamura, M, Miyazaki, T, Tanaka, Y, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparison of the structural changes in two cellobiohydrolases, CcCel6A and CcCel6C, from Coprinopsis cinerea - a tweezer-like motion in the structure of CcCel6C
Febs J., 279, 2012
|
|
2D73
 
 | | Crystal Structure Analysis of SusB | | Descriptor: | CALCIUM ION, alpha-glucosidase SusB | | Authors: | Kitamura, M, Yao, M. | | Deposit date: | 2005-11-15 | | Release date: | 2007-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
3W9A
 
 | | Crystal structure of the catalytic domain of the glycoside hydrolase family 131 protein from Coprinopsis cinerea | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Miyazaki, T, Tanaka, Y, Tamura, M, Yoshida, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-04-01 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the N-terminal domain of a glycoside hydrolase family 131 protein from Coprinopsis cinerea
Febs Lett., 587, 2013
|
|
3VV1
 
 | | Crystal Structure of Caenorhabditis elegans galectin LEC-6 | | Descriptor: | MAGNESIUM ION, Protein LEC-6, beta-D-galactopyranose-(1-4)-alpha-L-fucopyranose | | Authors: | Makyio, H, Takeuchi, T, Tamura, M, Nishiyama, K, Takahashi, H, Natsugari, H, Arata, Y, Kasai, K, Yamada, Y, Wakatsuki, S, Kato, R. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of preferential binding of fucose-containing saccharide by the Caenorhabditis elegans galectin LEC-6
Glycobiology, 23, 2013
|
|
8ITO
 
 | |
1WE5
 
 | | Crystal Structure of Alpha-Xylosidase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative family 31 glucosidase yicI | | Authors: | Ose, T, Kitamura, M, Okuyama, M, Mori, H, Kimura, A, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2004-05-24 | | Release date: | 2005-02-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Alpha-Xylosidase from Escherichia coli
TO BE PUBLISHED
|
|
5B37
 
 | | Crystal structure of L-tryptophan dehydrogenase from Nostoc punctiforme | | Descriptor: | Tryptophan dehydrogenase | | Authors: | Wakamatsu, T, Sakuraba, H, Kitamura, M, Hakumai, Y, Ohnishi, K, Ashiuchi, M, Ohshima, T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.
Appl. Environ. Microbiol., 83, 2017
|
|
5B3K
 
 | |
5B3L
 
 | |
1FLM
 
 | | DIMER OF FMN-BINDING PROTEIN FROM DESULFOVIBRIO VULGARIS (MIYAZAKI F) | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (FMN-BINDING PROTEIN) | | Authors: | Suto, K, Kawagoe, K, Shibata, N, Morimoto, K, Higuchi, Y, Kitamura, M, Nakaya, T, Yasuoka, N. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | How do the x-ray structure and the NMR structure of FMN-binding protein differ?
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7EG5
 
 | |
2CVC
 
 | | Crystal structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Hildenborough) | | Descriptor: | HEME C, High-molecular-weight cytochrome c precursor | | Authors: | Suto, K, Sato, M, Shibata, N, Kitamura, M, Morimoto, Y, Takayama, Y, Ozawa, K, Akutsu, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of High-Molecular Weight Cytochrome c
To be Published
|
|
2E84
 
 | | Crystal structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Miyazaki F) in the presence of zinc ion | | Descriptor: | High-molecular-weight cytochrome c, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Shibata, N, Suto, K, Sato, M, Morimoto, Y, Kitamura, M, Higuchi, Y. | | Deposit date: | 2007-01-17 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Miyazaki F)
To be Published
|
|
3A24
 
 | | Crystal structure of BT1871 retaining glycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, alpha-galactosidase | | Authors: | Okuyama, M, Kitamura, M, Hondoh, H, Tanaka, I, Yao, M. | | Deposit date: | 2009-04-28 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic mechanism of retaining alpha-galactosidase belonging to glycoside hydrolase family 97.
J.Mol.Biol., 392, 2009
|
|
2ZQ0
 
 | | Crystal structure of SusB complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase (Alpha-glucosidase SusB), CALCIUM ION | | Authors: | Yao, M, Tanaka, I, Kitamura, M. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
3A6R
 
 | | E13Q mutant of FMN-binding protein from Desulfovibrio vulgaris (Miyazaki F) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, FMN-binding protein | | Authors: | Nakanishi, T, Haruyama, Y, Inoue, H, Kitamura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effects of the disappearance of one charge on ultrafast fluorescence dynamics of the FMN binding protein.
J.Phys.Chem.B, 114, 2010
|
|
3W8Q
 
 | | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 (MEK1) | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Nakae, S, Kitamura, M, Shirai, T, Tada, T. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of mitogen-activated protein kinase kinase 1 in the DFG-out conformation.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3VY2
 
 | |
3VQT
 
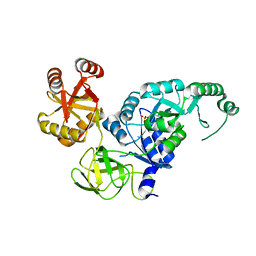 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
3VR1
 
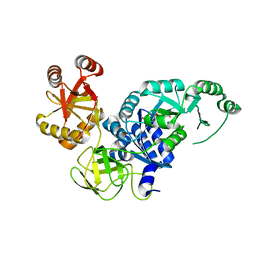 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-04-03 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
