5B1G
 
 | |
5B1E
 
 | |
5B1F
 
 | |
5B1D
 
 | |
2DCZ
 
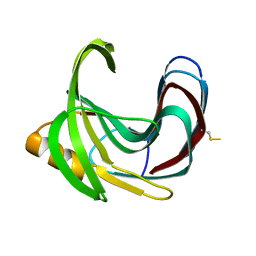 | | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase By Directed Evolution | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Endo-1,4-beta-xylanase A, SULFATE ION | | 著者 | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | 登録日 | 2006-01-18 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2E1C
 
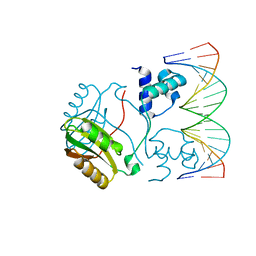 | | Structure of Putative HTH-type transcriptional regulator PH1519/DNA Complex | | 分子名称: | DNA (5'-D(*DAP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DTP*DGP*DAP*DAP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DCP*DAP*DCP*DT)-3'), Putative HTH-type transcriptional regulator PH1519 | | 著者 | Koike, H, Suzuki, M. | | 登録日 | 2006-10-24 | | 公開日 | 2007-12-04 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Feast/Famine Regulation by Transcription Factor FL11 for the Survival of the Hyperthermophilic Archaeon Pyrococcus OT3.
Structure, 15, 2007
|
|
2E5A
 
 | | Crystal Structure of Bovine Lipoyltransferase in Complex with Lipoyl-AMP | | 分子名称: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, ACETIC ACID, Lipoyltransferase 1, ... | | 著者 | Fujiwara, K, Hosaka, H, Matsuda, M, Suzuki, M, Nakagawa, A. | | 登録日 | 2006-12-19 | | 公開日 | 2007-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of bovine Lipoyltransferase in complex with lipoyl-AMP
J.Mol.Biol., 371, 2007
|
|
1ISP
 
 | | Crystal structure of Bacillus subtilis lipase at 1.3A resolution | | 分子名称: | GLYCEROL, lipase | | 著者 | Kawasaki, K, Kondo, H, Suzuki, M, Ohgiya, S, Tsuda, S. | | 登録日 | 2001-12-19 | | 公開日 | 2002-12-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5AZ1
 
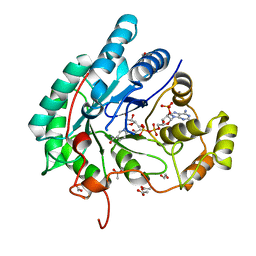 | | Crystal structure of aldo-keto reductase (AKR2E5) complexed with NADPH | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | 登録日 | 2015-09-15 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
1V5C
 
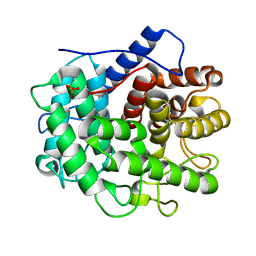 | | The crystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 | | 分子名称: | SULFATE ION, chitosanase | | 著者 | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | 登録日 | 2003-11-22 | | 公開日 | 2004-12-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
1V5D
 
 | | The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 | | 分子名称: | PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), chitosanase | | 著者 | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | 登録日 | 2003-11-22 | | 公開日 | 2004-12-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
1SQJ
 
 | | Crystal Structure Analysis of Oligoxyloglucan reducing-end-specific cellobiohydrolase (OXG-RCBH) | | 分子名称: | oligoxyloglucan reducing-end-specific cellobiohydrolase | | 著者 | Yaoi, K, Kondo, H, Noro, N, Suzuki, M, Tsuda, S, Mitsuishi, Y. | | 登録日 | 2004-03-19 | | 公開日 | 2004-07-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Tandem Repeat of a Seven-Bladed beta-Propeller Domain in Oligoxyloglucan Reducing-End-Specific Cellobiohydrolase
Structure, 12, 2004
|
|
7DDA
 
 | | Envelope protein VP37 a crystal structure from White Spot Syndrome Virus | | 分子名称: | Envelope protein, SULFATE ION | | 著者 | Somsoros, W, Sangawa, T, Takebe, K, Attarataya, J, Suzuki, M, Khunrae, P. | | 登録日 | 2020-10-28 | | 公開日 | 2021-06-23 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structure of the C-terminal domain of envelope protein VP37 from white spot syndrome virus reveals sulphate binding sites responsible for heparin binding.
J.Gen.Virol., 102, 2021
|
|
2D05
 
 | | Chitosanase From Bacillus circulans mutant K218P | | 分子名称: | Chitosanase, SULFATE ION | | 著者 | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | 登録日 | 2005-07-25 | | 公開日 | 2005-12-06 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
5B21
 
 | |
5AZ0
 
 | | Crystal structure of aldo-keto reductase (AKR2E5) of the silkworm, Bombyx mori | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | 登録日 | 2015-09-15 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5B22
 
 | | Dimer structure of murine Nectin-3 D1D2 | | 分子名称: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | 登録日 | 2015-12-28 | | 公開日 | 2016-12-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
2E0Z
 
 | | Crystal structure of virus-like particle from Pyrococcus furiosus | | 分子名称: | Virus-like particle | | 著者 | Akita, F, Chong, K.T, Tanaka, H, Yamashita, E, Miyazaki, N, Nakaishi, Y, Namba, K, Ono, Y, Suzuki, M, Tsukihara, T, Nakagawa, A. | | 登録日 | 2006-10-16 | | 公開日 | 2007-04-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses
J.Mol.Biol., 368, 2007
|
|
1RI7
 
 | |
5WR9
 
 | | Crystal structure of hen egg-white lysozyme | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | 登録日 | 2016-12-01 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WR8
 
 | | Thaumatin structure determined by SACLA at 1.55 Angstrom | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin I | | 著者 | Masuda, T, Suzuki, M, Inoue, S, Sugahara, M. | | 登録日 | 2016-12-01 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WRB
 
 | | Crystal structure of hen egg-white lysozyme | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | 登録日 | 2016-12-01 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.013 Å) | | 主引用文献 | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WRC
 
 | | Crystal structure of proteinase K from Engyodontium album | | 分子名称: | NITRATE ION, PRASEODYMIUM ION, Proteinase K | | 著者 | Sugahara, M, Nakane, T, Suzuki, M, Masuda, T, Inoue, S, Numata, K. | | 登録日 | 2016-12-01 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5WRA
 
 | | Crystal structure of hen egg-white lysozyme | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | 登録日 | 2016-12-01 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5X9M
 
 | | Structure of hyper-sweet thaumatin (D21N) | | 分子名称: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | 著者 | Masuda, T, Okubo, K, Sugahara, M, Suzuki, M, Mikami, B. | | 登録日 | 2017-03-08 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.93 Å) | | 主引用文献 | Subatomic structure of hyper-sweet thaumatin D21N mutant reveals the importance of flexible conformations for enhanced sweetness.
Biochimie, 157, 2019
|
|
