5GR0
 
 | |
5GQU
 
 | |
5GR6
 
 | |
5GR3
 
 | |
5GQV
 
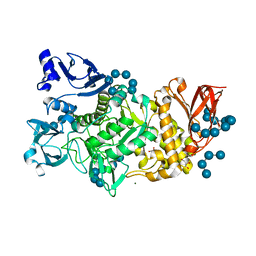 | |
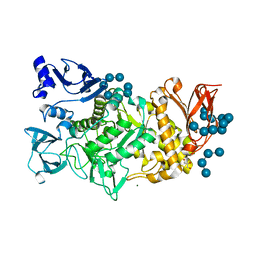
5GR4
 
 | |
5GR1
 
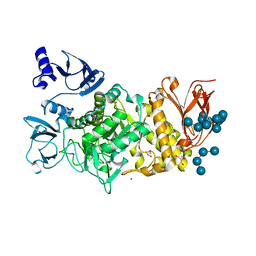 | |
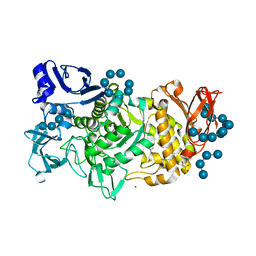
5GQY
 
 | |
5GR2
 
 | |
5GR5
 
 | |
5GQW
 
 | |
5GQX
 
 | |
5GQZ
 
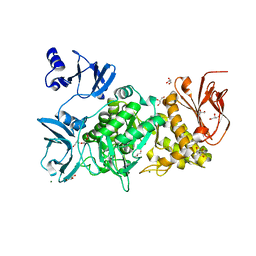 | |
6KLF
 
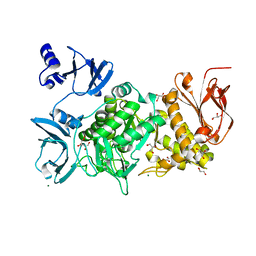 | |
1PHS
 
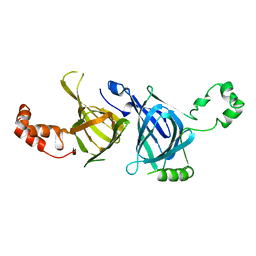 | | THE THREE-DIMENSIONAL STRUCTURE OF THE SEED STORAGE PROTEIN PHASEOLIN AT 3 ANGSTROMS RESOLUTION | | 分子名称: | PHASEOLIN, BETA-TYPE PRECURSOR | | 著者 | Lawrence, M.C, Suzuki, E, Varghese, J.N, Davis, P.C, Vandonkelaar, A, Tulloch, P.A, Colman, P.M. | | 登録日 | 1990-03-21 | | 公開日 | 1990-10-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The three-dimensional structure of the seed storage protein phaseolin at 3 A resolution.
EMBO J., 9, 1990
|
|
2DPF
 
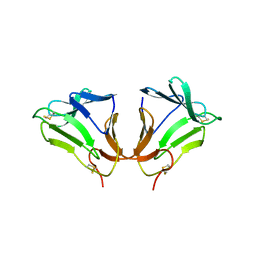 | | Crystal Structure of curculin1 homodimer | | 分子名称: | Curculin, SULFATE ION | | 著者 | Kurimoto, E, Suzuki, M, Amemiya, E, Yamaguchi, Y, Nirasawa, S, Shimba, N, Xu, N, Kashiwagi, T, Kawai, M, Suzuki, E, Kato, K. | | 登録日 | 2006-05-11 | | 公開日 | 2007-05-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Curculin Exhibits Sweet-tasting and Taste-modifying Activities through Its Distinct Molecular Surfaces.
J.Biol.Chem., 282, 2007
|
|
1BXV
 
 | | REDUCED PLASTOCYANIN FROM SYNECHOCOCCUS SP. | | 分子名称: | COPPER (II) ION, PLASTOCYANIN | | 著者 | Inoue, T, Sugawara, H, Hamanaka, S, Tsukui, H, Suzuki, E, Kohzuma, T, Kai, Y. | | 登録日 | 1998-10-09 | | 公開日 | 1999-06-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure determinations of oxidized and reduced plastocyanin from the cyanobacterium Synechococcus sp. PCC 7942.
Biochemistry, 38, 1999
|
|
1BXU
 
 | | OXIDIZED PLASTOCYANIN FROM SYNECHOCOCCUS SP. | | 分子名称: | COPPER (II) ION, PLASTOCYANIN | | 著者 | Inoue, T, Sugawara, H, Hamanaka, S, Tsukui, H, Suzuki, E, Kohzuma, T, Kai, Y. | | 登録日 | 1998-10-09 | | 公開日 | 1999-06-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure determinations of oxidized and reduced plastocyanin from the cyanobacterium Synechococcus sp. PCC 7942.
Biochemistry, 38, 1999
|
|
4HE7
 
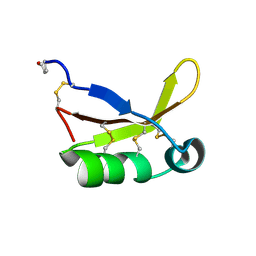 | | Crystal Structure of Brazzein | | 分子名称: | Defensin-like protein, SODIUM ION | | 著者 | Nagata, K, Hongo, N, Kameda, Y, Yamamura, A, Sasaki, H, Lee, W.C, Ishikawa, K, Suzuki, E, Tanokura, M. | | 登録日 | 2012-10-03 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2KSV
 
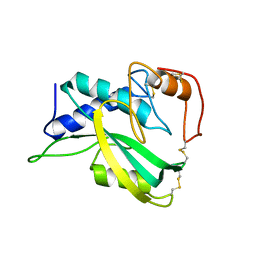 | | The NMR structure of protein-glutaminase from Chryseobacterium proteolyticum | | 分子名称: | Protein-glutaminase | | 著者 | Kumeta, H, Miwa, N, Ogura, K, Kai, Y, Mizukoshi, T, Shimba, N, Suzuki, E, Inagaki, F. | | 登録日 | 2010-01-14 | | 公開日 | 2010-02-16 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR structure of protein-glutaminase from Chryseobacterium proteolyticum.
J.Biomol.Nmr, 46, 2010
|
|
1EOI
 
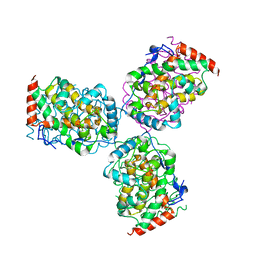 | | CRYSTAL STRUCTURE OF ACID PHOSPHATASE FROM ESCHERICHIA BLATTAE COMPLEXED WITH THE TRANSITION STATE ANALOG MOLYBDATE | | 分子名称: | ACID PHOSPHATASE, MOLYBDATE ION | | 著者 | Ishikawa, K, Mihara, Y, Gondoh, K, Suzuki, E, Asano, Y. | | 登録日 | 2000-03-23 | | 公開日 | 2001-03-23 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate.
EMBO J., 19, 2000
|
|
1DET
 
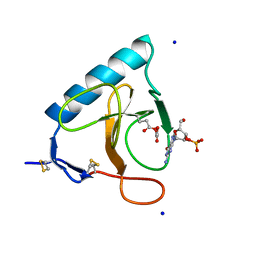 | | RIBONUCLEASE T1 CARBOXYMETHYLATED AT GLU 58 IN COMPLEX WITH 2'GMP | | 分子名称: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | 著者 | Ishikawa, K, Suzuki, E, Tanokura, M, Takahashi, K. | | 登録日 | 1996-02-20 | | 公開日 | 1996-07-11 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of ribonuclease T1 carboxymethylated at Glu58 in complex with 2'-GMP.
Biochemistry, 35, 1996
|
|
1D2T
 
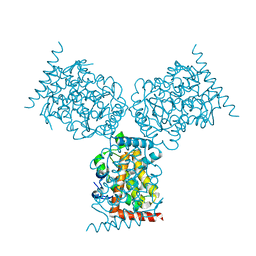 | | CRYSTAL STRUCTURE OF ACID PHOSPHATASE FROM ESCHERICHIA BLATTAE | | 分子名称: | ACID PHOSPHATASE, SULFATE ION | | 著者 | Ishikawa, K, Mihara, Y, Gondoh, K, Suzuki, E, Asano, Y. | | 登録日 | 1999-09-28 | | 公開日 | 2000-12-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate.
EMBO J., 19, 2000
|
|
1IW8
 
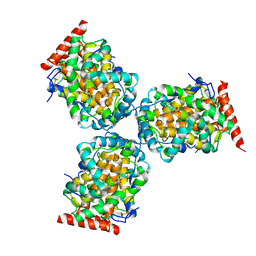 | | Crystal Structure of a mutant of acid phosphatase from Escherichia blattae (G74D/I153T) | | 分子名称: | SULFATE ION, acid phosphatase | | 著者 | Ishikawa, K, Mihara, Y, Shimba, N, Ohtsu, N, Kawasaki, H, Suzuki, E, Asano, Y. | | 登録日 | 2002-04-22 | | 公開日 | 2002-09-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Enhancement of nucleoside phosphorylation activity in an acid phosphatase
PROTEIN ENG., 15, 2002
|
|
1IYY
 
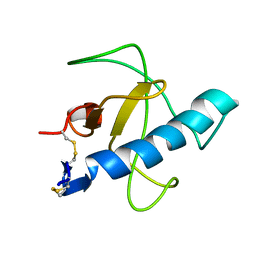 | | NMR STRUCTURE OF Gln25-RIBONUCLEASE T1, 24 STRUCTURES | | 分子名称: | RIBONUCLEASE T1 | | 著者 | Hatano, K, Kojima, M, Suzuki, E, Tanokura, M, Takahashi, K. | | 登録日 | 2002-09-12 | | 公開日 | 2003-10-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Determination of the NMR structure of Gln25-ribonuclease T1.
Biol. Chem., 384, 2003
|
|
