4MTP
 
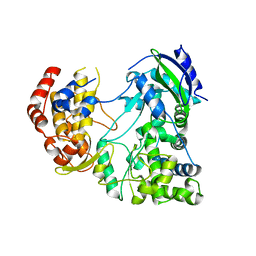 | | RdRp from Japanesese Encephalitis Virus | | 分子名称: | RNA dependent RNA polymerase, ZINC ION | | 著者 | Surana, P, Nair, D.T. | | 登録日 | 2013-09-20 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state.
Nucleic Acids Res., 42, 2014
|
|
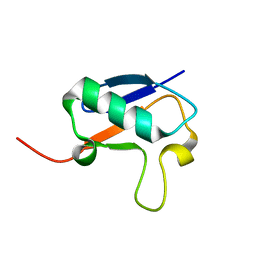
2NBD
 
 | |
2NBE
 
 | |
4HDH
 
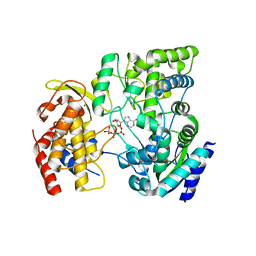 | | Crystal Structure of viral RdRp in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Polyprotein, ZINC ION | | 著者 | Surana, P, Nair, D.T. | | 登録日 | 2012-10-02 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state.
Nucleic Acids Res., 42, 2014
|
|
4HDG
 
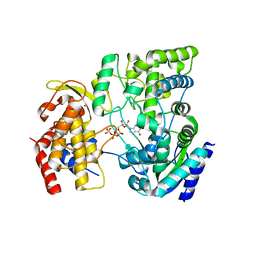 | | Crystal Structure of viral RdRp in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Polyprotein, ZINC ION | | 著者 | Surana, P, Nair, D.T. | | 登録日 | 2012-10-02 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state.
Nucleic Acids Res., 42, 2014
|
|
5YCU
 
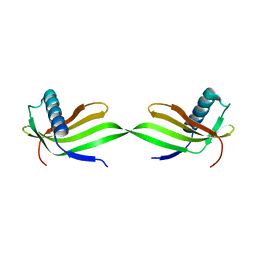 | | Domain swapped dimer of engineered hairpin loop1 mutant in Single-chain Monellin | | 分子名称: | Single chain monellin | | 著者 | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | 登録日 | 2017-09-08 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
5YCW
 
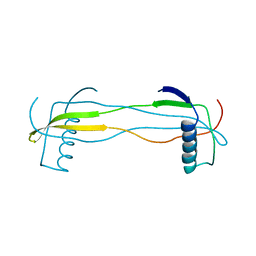 | | Double domain swapped dimer of engineered hairpin loop1 and loop3 mutant in Single-chain Monellin | | 分子名称: | single chain monellin | | 著者 | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | 登録日 | 2017-09-08 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.285 Å) | | 主引用文献 | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
5YCT
 
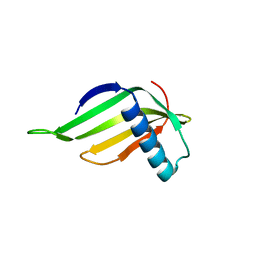 | | Engineered hairpin loop3 mutant monomer in Single-chain Monellin | | 分子名称: | Single chain Monellin | | 著者 | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | 登録日 | 2017-09-08 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
5XFU
 
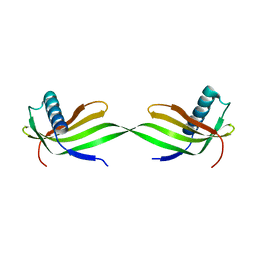 | | Domain swapped dimer crystal structure of loop1 deletion mutant in Single-chain Monellin | | 分子名称: | Monellin chain B,Monellin chain A | | 著者 | Surana, P, Nandwani, N, Udgaonkar, J, Gosavi, S, Das, R. | | 登録日 | 2017-04-11 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.611 Å) | | 主引用文献 | Amino-acid composition after loop deletion drives domain swapping
Protein Sci., 26, 2017
|
|
5XQM
 
 | |
