5K9H
 
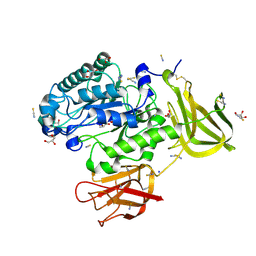 | | Crystal structure of a glycoside hydrolase 29 family member from an unknown rumen bacterium | | Descriptor: | 0940_GH29, GLYCEROL, SODIUM ION, ... | | Authors: | Summers, E.L, Arcus, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The structure of a glycoside hydrolase 29 family member from a rumen bacterium reveals unique, dual carbohydrate-binding domains.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
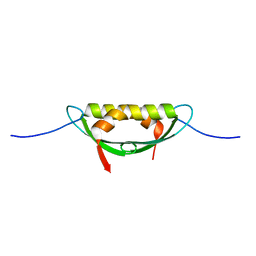
4E1P
 
 | |
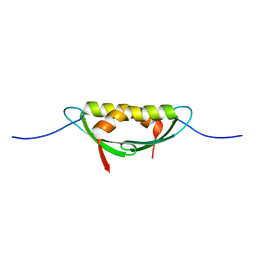
4E1R
 
 | |
7RA4
 
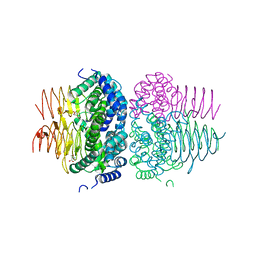 | | Crystal structure of Neisseria gonorrhoeae serine acetyltransferase (CysE) in complex with serine | | Descriptor: | SERINE, Serine acetyltransferase | | Authors: | Hicks, J.L, Oldham, K.E, Prentice, E.J, Summers, E.L. | | Deposit date: | 2021-06-30 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serine acetyltransferase from Neisseria gonorrhoeae; structural and biochemical basis of inhibition.
Biochem.J., 479, 2022
|
|
6WYE
 
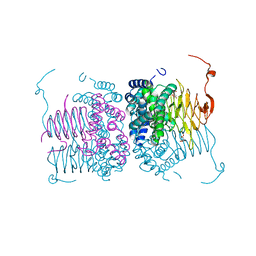 | | Crystal structure of Neisseria gonorrhoeae serine acetyltransferase (CysE) | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SODIUM ION, Serine acetyltransferase | | Authors: | Hicks, J.L, Oldham, K.E, Summers, E.L, Prentice, E.J. | | Deposit date: | 2020-05-12 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Serine acetyltransferase from Neisseria gonorrhoeae; structural and biochemical basis of inhibition.
Biochem.J., 479, 2022
|
|
5U7V
 
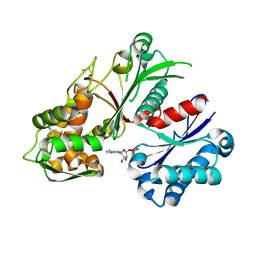 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Trifolium repens in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Apyrase | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
5U7P
 
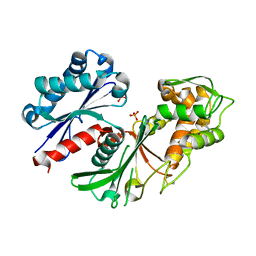 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Trifolium repens | | Descriptor: | Apyrase, PHOSPHATE ION | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
5U7W
 
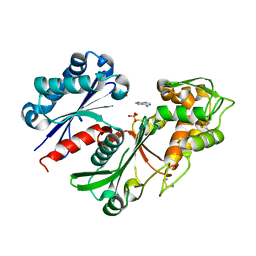 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Trifolium repens in complex with adenine and phosphate | | Descriptor: | ADENINE, Apyrase, PHOSPHATE ION | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
5U7X
 
 | | Crystal structure of a nucleoside triphosphate diphosphohydrolase (NTPDase) from the legume Vigna unguiculata subsp. cylindrica (Dolichos biflorus) in complex with phosphate and manganese | | Descriptor: | MANGANESE (II) ION, Nod factor binding lectin-nucleotide phosphohydrolase, PHOSPHATE ION | | Authors: | Cumming, M.H, Summers, E.L, Oulavallickal, T, Roberts, N, Arcus, V.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures and kinetics for plant nucleoside triphosphate diphosphohydrolases support a domain motion catalytic mechanism.
Protein Sci., 26, 2017
|
|
