1ZX9
 
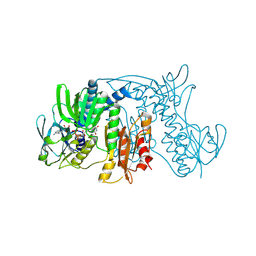 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mercuric reductase | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-06-07 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
1ZK7
 
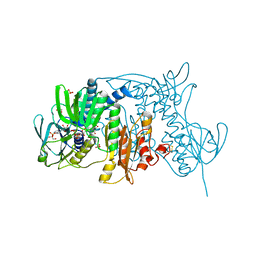 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase, ... | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-05-02 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
1S6L
 
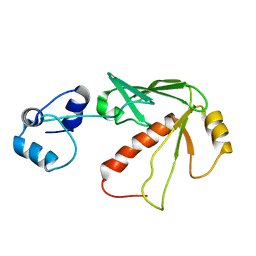 | | Solution structure of MerB, the Organomercurial Lyase involved in the bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Di Lello, P, Benison, G.C, Valafar, H, Pitts, K.E, Summers, A.O, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-01-25 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system.
Biochemistry, 43, 2004
|
|
