1GSO
 
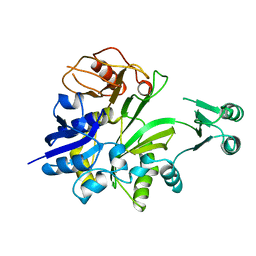 | |
1N0O
 
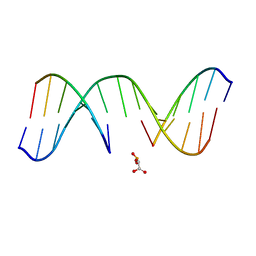 | | NMR Structure of d(CCAAGGXCTTGGG), X is a 3'-phosphoglycolate, 5'-phosphate gapped lesion, 10 structures | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*GP*G)-3', 5'-D(*CP*CP*CP*AP*AP*GP*GP*CP*CP*TP*TP*GP*G)-3', ... | | Authors: | Junker, H.-D, Hoehn, S.T, Bunt, R.C, Marathius, V, Chen, J, Turner, C.J, Stubbe, J. | | Deposit date: | 2002-10-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Characterization, and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Nucleic Acids Res., 30, 2002
|
|
1JK0
 
 | | Ribonucleotide reductase Y2Y4 heterodimer | | Descriptor: | ZINC ION, ribonucleoside-diphosphate reductase small chain 1, ribonucleoside-diphosphate reductase small chain 2 | | Authors: | Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2001-07-10 | | Release date: | 2001-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the yeast ribonucleotide reductase Y2Y4 heterodimer.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1N0K
 
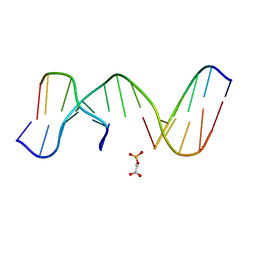 | | NMR Structure of duplex DNA d(CCAAGGXCTTGGG), X is a 3' phosphoglycolate, 5'phosphate gapped lesion | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*GP*G)-3', 5'-D(*CP*CP*CP*AP*AP*GP*GP*CP*CP*TP*TP*GP*G)-3', ... | | Authors: | Junker, H.-D, Hoehn, S.T, Bunt, R.C, Marathius, V, Chen, J, Turner, C.J, Stubbe, J. | | Deposit date: | 2002-10-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Characterization and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Nucleic Acids Res., 30, 2002
|
|
1T3T
 
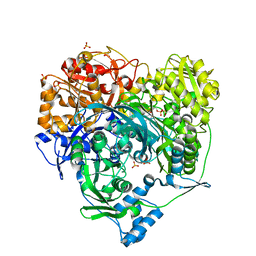 | | Structure of Formylglycinamide synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | Authors: | Ealick, S.E, Anand, R, Hoskin, A.A, Stubbe, J. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Domain Organization of Salmonella typhimurium Formylglycinamide Ribonucleotide Amidotransferase Revealed by X-ray crystallography
Biochemistry, 43, 2004
|
|
1L1L
 
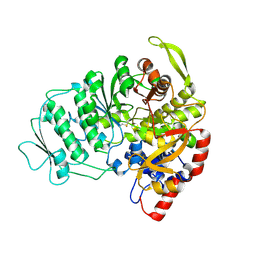 | | CRYSTAL STRUCTURE OF B-12 DEPENDENT (CLASS II) RIBONUCLEOTIDE REDUCTASE | | Descriptor: | RIBONUCLEOSIDE TRIPHOSPHATE REDUCTASE | | Authors: | Sintchak, M.D, Arjara, G, Kellogg, B.A, Stubbe, J, Drennan, C.L. | | Deposit date: | 2002-02-18 | | Release date: | 2002-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer.
Nat.Struct.Biol., 9, 2002
|
|
1SMS
 
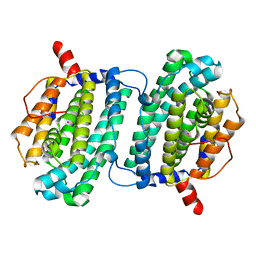 | | Structure of the Ribonucleotide Reductase Rnr4 Homodimer from Saccharomyces cerevisiae | | Descriptor: | MERCURY (II) ION, Ribonucleoside-diphosphate reductase small chain 2 | | Authors: | Sommerhalter, M, Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers.
Biochemistry, 43, 2004
|
|
1SMQ
 
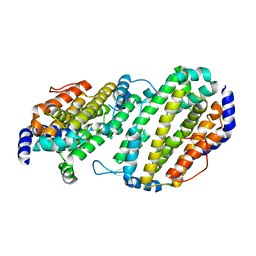 | | Structure of the Ribonucleotide Reductase Rnr2 Homodimer from Saccharomyces cerevisiae | | Descriptor: | Ribonucleoside-diphosphate reductase small chain 1 | | Authors: | Sommerhalter, M, Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers.
Biochemistry, 43, 2004
|
|
4DR0
 
 | | Crystal structure of Bacillus subtilis dimanganese(II) NrdF | | Descriptor: | MANGANESE (II) ION, Ribonucleoside-diphosphate reductase subunit beta, SULFATE ION | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2012-02-16 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Dimanganese(II) Site of Bacillus subtilis Class Ib Ribonucleotide Reductase.
Biochemistry, 51, 2012
|
|
1T4A
 
 | | Structure of B. Subtilis PurS C2 Crystal Form | | Descriptor: | PurS | | Authors: | Anand, R, Hoskins, A.A, Bennett, E.M, Sintchak, M.D, Stubbe, J, Ealick, S.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A model for the Bacillus subtilis formylglycinamide ribonucleotide amidotransferase multiprotein complex.
Biochemistry, 43, 2004
|
|
1TWJ
 
 | |
