4ZQX
 
 | | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Ginn, H.M, Brewster, A.S, Hattne, J, Evans, G, Wagner, A, Grimes, J, Sauter, N.K, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-05-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7Q0I
 
 | | Crystal structure of the N-terminal domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-44 and Beta-54 Fabs | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-44 Fab heavy chain, Beta-44 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0G
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-49 and FI-3A Fabs | | Descriptor: | Beta-49 Fab heavy chain, Beta-49 Fab light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0H
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-50 and Beta-54 | | Descriptor: | Beta-50 Fab heavy chain, Beta-50 Fab light chain, Beta-54 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0A
 
 | | SARS-CoV-2 Spike ectodomain with Fab FI3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI3A fab Light chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
1OOP
 
 | | The Crystal Structure of Swine Vesicular Disease Virus | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Knowles, N.J, Newman, J.W.I, Wilsden, G, Rao, Z, King, A.M.Q, Stuart, D.I. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Swine Vesicular Disease Virus and Implications for Host Adaptation
J.Virol., 77, 2003
|
|
1W46
 
 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W48
 
 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W47
 
 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
5A99
 
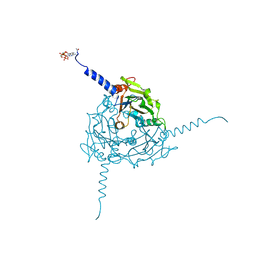 | | Crystal structure of Operophtera brumata CPV19 polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
1W8X
 
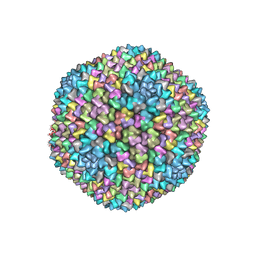 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
5A8S
 
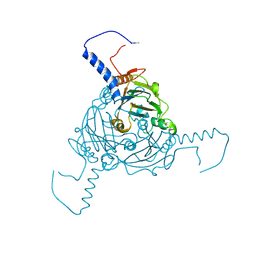 | | Crystal structure of Antheraea mylitta CPV4 polyhedra type 1 | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5AC9
 
 | | Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, PerezMartin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-Based Energetics of Protein Interfaces Guide Foot-and-Mouth Disease Vaccine Design
Nat.Struct.Mol.Biol., 22, 2015
|
|
1W49
 
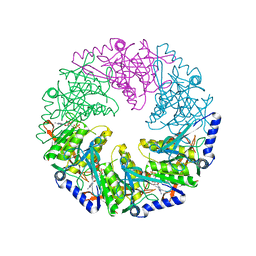 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP and Mg | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W4B
 
 | | P4 protein from PHI12 in complex with product (AMPcPP Mg 22C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1OGA
 
 | | A structural basis for immunodominant human T-cell receptor recognition. | | Descriptor: | BETA-2-MICROGLOBULIN, GILGFVFTL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B.E, McMichael, A.J, Bell, J.I, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2003-04-28 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Structural Basis for Immunodominant Human T Cell Receptor Recognition
Nat.Immunol., 4, 2003
|
|
1WAC
 
 | | Back-priming mode of Phi6 RNA-dependent RNA polymerase | | Descriptor: | P2 PROTEIN | | Authors: | Laurila, M.R.L, Salgado, P.S, Stuart, D.I, Grimes, J.M, Bamford, D.H. | | Deposit date: | 2004-10-26 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Back-Priming Mode of Phi6 RNA-Dependent RNA Polymerase
J.Gen.Virol., 86, 2005
|
|
1W9Z
 
 | | Structure of Bannavirus VP9 | | Descriptor: | VP9 | | Authors: | Jaafar, F.M, Attoui, H, Bahar, M.W, Siebold, C, Sutton, G, Mertens, P.P.C, Micco, P, Stuart, D.I, Grimes, J.M, Lamballerie, X. | | Deposit date: | 2004-10-21 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The Structure and Function of the Outer Coat Protein Vp9 of Banna Virus
Structure, 13, 2005
|
|
1W4C
 
 | | P4 protein from Bacteriophage PHI12 apo state | | Descriptor: | NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
5A9C
 
 | | Crystal structure of Antheraea mylitta CPV4 polyhedra base domain deleted mutant | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A98
 
 | | Crystal structure of Trichoplusia ni CPV15 polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A96
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
1OLZ
 
 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
5ABJ
 
 | | Structure of Coxsackievirus A16 in complex with GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
