6Y5X
 
 | |
6Y6A
 
 | | Structure of Finch Polyomavirus VP1 in complex with 2-O-Methyl-5-N-acetyl-alpha-D-neuraminic acid | | Descriptor: | CHLORIDE ION, Capsid protein VP1, MAGNESIUM ION, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y66
 
 | | Structure of Goose Hemorrhagic Polyomavirus VP1 in complex with 2-O-Methyl-5-N-acetyl-alpha-D-neuraminic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, Capsid protein VP1, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
3O8E
 
 | | Structure of extracelllar portion of CD46 in complex with Adenovirus type 11 knob | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DITHIANE DIOL, Fiber 36.1 kDa protein, ... | | Authors: | Persson, B.D, Schmitz, N.B, Casasnovas, J.M, Stehle, T. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens.
Plos Pathog., 6, 2010
|
|
7A9C
 
 | |
5C1J
 
 | |
7AJP
 
 | | Crystal Structure of Human Adenovirus 56 Fiber Knob | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLYCEROL, ... | | Authors: | Strebl, M, Mindler, K, Stehle, T. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.382018 Å) | | Cite: | Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3PH5
 
 | |
3PH6
 
 | |
5CPU
 
 | | Crystal structure of murine polyomavirus PTA strain VP1 | | Descriptor: | GLYCEROL, VP1 | | Authors: | Liaci, A.M, Buch, M.H.C, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
5CQ0
 
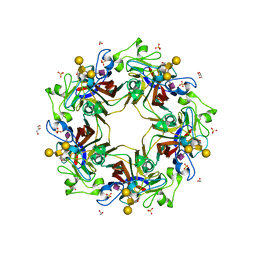 | | Crystal structure of murine polyomavirus RA strain VP1 in complex with the GD1a glycan | | Descriptor: | Capsid protein VP1, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Buch, M.H.C, Liaci, A.M, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
5CPX
 
 | | Crystal structure of murine polyomavirus PTA strain VP1 in complex with the DSLNT glycan | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Buch, M.H.C, Liaci, A.M, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
7B27
 
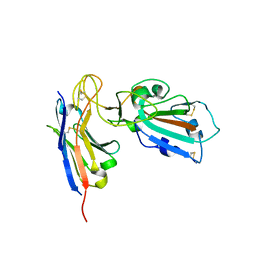 | |
6G47
 
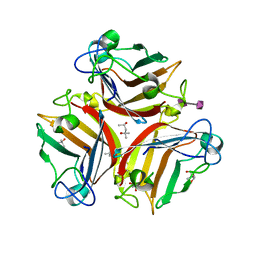 | | Crystal Structure of Human Adenovirus 52 Short Fiber Knob in Complex with alpha-(2,8)-Trisialic Acid (DP3) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Liaci, A.M, Stehle, T. | | Deposit date: | 2018-03-26 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Polysialic acid is a cellular receptor for human adenovirus 52.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5CPW
 
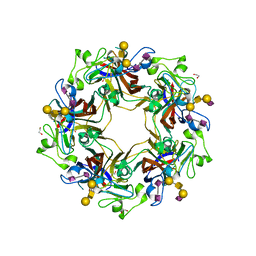 | | Crystal structure of murine polyomavirus PTA strain VP1 in complex with the GT1a glycan | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Liaci, A.M, Buch, M.H.C, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
6ZS0
 
 | | Crystal structure of 5-dimethylallyltryptophan synthase from Streptomyces coelicolor | | Descriptor: | DI(HYDROXYETHYL)ETHER, DMATS type aromatic prenyltransferase | | Authors: | Ostertag, E, Broger, K, Stehle, T, Zocher, G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reprogramming Substrate and Catalytic Promiscuity of Tryptophan Prenyltransferases.
J.Mol.Biol., 433, 2020
|
|
6GS2
 
 | | Crystal Structure of the GatD/MurT Enzyme Complex from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, SA1707 protein, ... | | Authors: | Muckenfuss, L.M, Noeldeke, E.R, Niemann, V, Zocher, G, Stehle, T. | | Deposit date: | 2018-06-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cell wall peptidoglycan amidation by the GatD/MurT complex of Staphylococcus aureus.
Sci Rep, 8, 2018
|
|
6ZRX
 
 | | Crystal structure of 6-dimethylallyltryptophan synthase from Micromonospora olivasterospora in complex with DMASPP and Trp | | Descriptor: | DI(HYDROXYETHYL)ETHER, DMATS type aromatic prenyltransferase, S-(3-methylbut-2-en-1-yl) trihydrogen thiodiphosphate, ... | | Authors: | Ostertag, E, Stehle, T, Zocher, G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reprogramming Substrate and Catalytic Promiscuity of Tryptophan Prenyltransferases.
J.Mol.Biol., 433, 2020
|
|
6ZRY
 
 | | 6-dimethylallyl tryptophan synthase | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DMATS type aromatic prenyltransferase, ... | | Authors: | Ostertag, E, Stehle, T, Zocher, G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Reprogramming Substrate and Catalytic Promiscuity of Tryptophan Prenyltransferases.
J.Mol.Biol., 433, 2020
|
|
6ZLZ
 
 | |
6ZML
 
 | | CryoEM Structure of Merkel Cell Polyomavirus Virus-like Particle | | Descriptor: | Capsid protein VP1 | | Authors: | Bayer, N.J, Januliene, D, Stehle, T, Moeller, A, Blaum, B.S. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Merkel Cell Polyomavirus Capsid and Interaction with Its Glycosaminoglycan Attachment Receptor.
J.Virol., 94, 2020
|
|
6H5E
 
 | | Crystal Structure of the GatD/MurT Enzyme Complex from Staphylococcus aureus with bound AMPPNP | | Descriptor: | DI(HYDROXYETHYL)ETHER, DUF1727 domain-containing protein, GLYCEROL, ... | | Authors: | Noeldeke, E.R, Niemann, V, Stoerk, E, Stehle, T. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.139 Å) | | Cite: | Structural basis of cell wall peptidoglycan amidation by the GatD/MurT complex of Staphylococcus aureus.
Sci Rep, 8, 2018
|
|
5EMI
 
 | | N-acetylmuramoyl-L-alanine amidase AmiC2 of Nostoc punctiforme | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cell wall hydrolase/autolysin, ... | | Authors: | Buettner, F.M, Stehle, T. | | Deposit date: | 2015-11-06 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Enabling cell-cell communication via nanopore formation: structure, function and localization of the unique cell wall amidase AmiC2 of Nostoc punctiforme.
Febs J., 283, 2016
|
|
5CPY
 
 | | Crystal structure of murine polyomavirus PTA strain VP1 in complex with the GD1a glycan | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, VP1 | | Authors: | Liaci, A.M, Buch, M.H.C, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
5CPZ
 
 | | Crystal structure of murine polyomavirus RA strain VP1 in complex with the GT1a glycan | | Descriptor: | Capsid protein VP1, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Buch, M.H.C, Liaci, A.M, Neu, U, Stehle, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
