3UFB
 
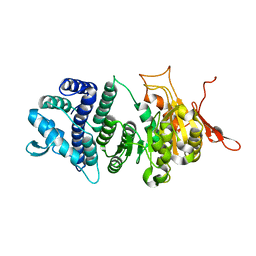 | | Crystal structure of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016 | | Descriptor: | Type I restriction-modification system methyltransferase subunit | | Authors: | Park, S.Y, Lee, H.J, Sun, J, Nishi, K, Song, J.M, Kim, J.S. | | Deposit date: | 2011-11-01 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of a modification subunit of a putative type I restriction enzyme from Vibrio vulnificus YJ016
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5KBW
 
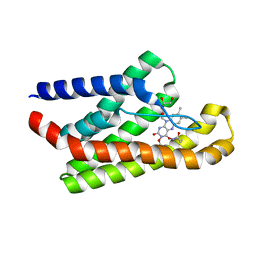 | |
5KC4
 
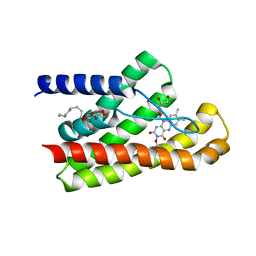 | | Structure of TmRibU, orthorhombic crystal form | | Descriptor: | RIBOFLAVIN, Riboflavin transporter RibU, nonyl beta-D-glucopyranoside | | Authors: | Karpowich, N.K, Wang, D.N, Song, J.M. | | Deposit date: | 2016-06-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Aromatic Cap Seals the Substrate Binding Site in an ECF-Type S Subunit for Riboflavin.
J.Mol.Biol., 428, 2016
|
|
5KC0
 
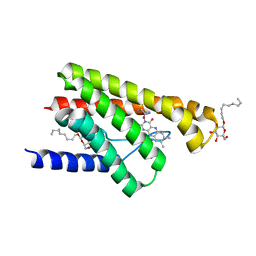 | | Crystal structure of TmRibU, hexagonal crystal form | | Descriptor: | RIBOFLAVIN, Riboflavin transporter RibU, nonyl beta-D-glucopyranoside | | Authors: | Karpowich, N.K, Wang, D.N, Song, J.M. | | Deposit date: | 2016-06-03 | | Release date: | 2016-06-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.2001 Å) | | Cite: | An Aromatic Cap Seals the Substrate Binding Site in an ECF-Type S Subunit for Riboflavin.
J.Mol.Biol., 428, 2016
|
|
7LO7
 
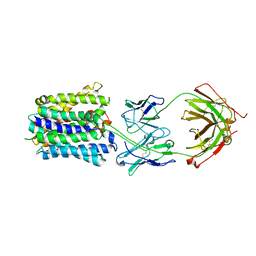 | | NorA in complex with Fab25 | | Descriptor: | Fab25 Heavy Chain, Fab25 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
7LO8
 
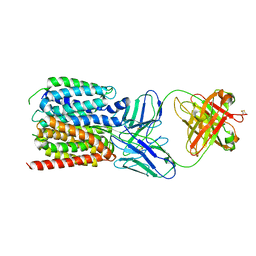 | | NorA in complex with Fab36 | | Descriptor: | Fab36 Heavy Chain, Fab36 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
7T9F
 
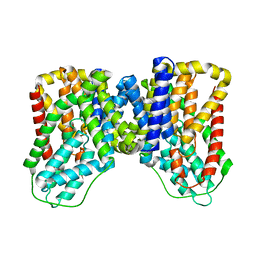 | | Structure of VcINDY-apo | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Song, J.M, Wang, D.N. | | Deposit date: | 2021-12-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of ion - substrate coupling in the Na + -dependent dicarboxylate transporter VcINDY.
Nat Commun, 13, 2022
|
|
7T9G
 
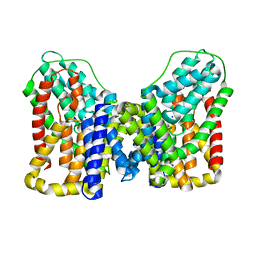 | | Structure of VcINDY-Na+ | | Descriptor: | DASS family sodium-coupled anion symporter, SODIUM ION | | Authors: | Sauer, D.B, Marden, J.J, Song, J.M, Wang, D.N. | | Deposit date: | 2021-12-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis of ion - substrate coupling in the Na + -dependent dicarboxylate transporter VcINDY.
Nat Commun, 13, 2022
|
|
6WW5
 
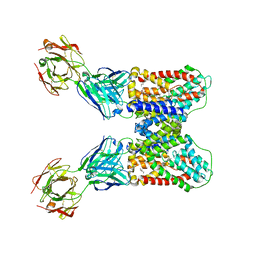 | | Structure of VcINDY-Na-Fab84 in nanodisc | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, DASS family sodium-coupled anion symporter, Fab84 Heavy Chain, ... | | Authors: | Sauer, D.B, Marden, J, Song, J.M, Koide, A, Koide, S, Wang, D.N. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU1
 
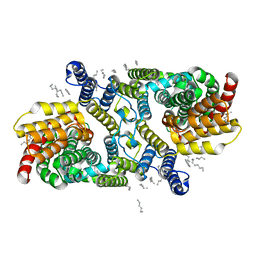 | | Structure of apo LaINDY | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N.C, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU2
 
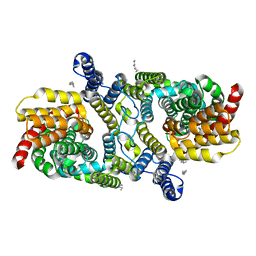 | | Structure of the LaINDY-malate complex | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WTW
 
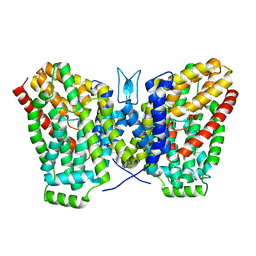 | | Structure of LaINDY crystallized in the presence of alpha-ketoglutarate and malate | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Cocco, N, Marden, J.J, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU4
 
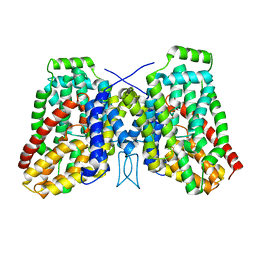 | | Structure of the LaINDY-alpha-ketoglutarate complex | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU3
 
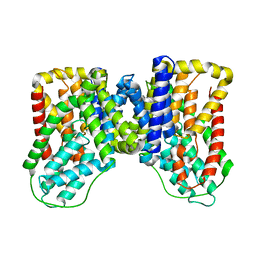 | |
4ZIR
 
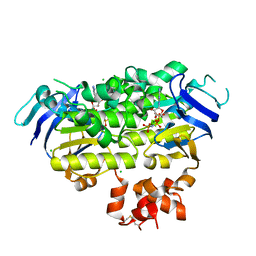 | | Crystal structure of EcfAA' heterodimer bound to AMPPNP | | Descriptor: | CHLORIDE ION, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Karpowich, N.K, Cocco, N, Song, J.M, Wang, D.N. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATP binding drives substrate capture in an ECF transporter by a release-and-catch mechanism.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3OX4
 
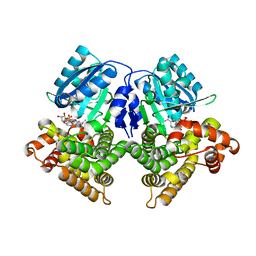 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 complexed with NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
3OWO
 
 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
