4FBC
 
 | | Structure of mutant RIP from barley seeds in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | 著者 | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | 登録日 | 2012-05-22 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4HAN
 
 | | Crystal structure of Galectin 8 with NDP52 peptide | | 分子名称: | Calcium-binding and coiled-coil domain-containing protein 2, DI(HYDROXYETHYL)ETHER, Galectin-8, ... | | 著者 | Kim, B.-W, Hong, S.B, Kim, J.H, Kwon, D.H, Song, H.K. | | 登録日 | 2012-09-27 | | 公開日 | 2013-03-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.551 Å) | | 主引用文献 | Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8.
Nat Commun, 4, 2013
|
|
4GQW
 
 | |
4FB9
 
 | | Structure of mutant RIP from barley seeds | | 分子名称: | Protein synthesis inhibitor I | | 著者 | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | 登録日 | 2012-05-22 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4GQY
 
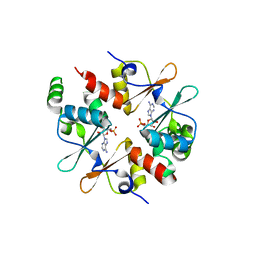 | | Crystal structure of CBSX2 in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, CBS domain-containing protein CBSX2, chloroplastic | | 著者 | Jeong, B.C, Song, H.K. | | 登録日 | 2012-08-24 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.193 Å) | | 主引用文献 | Change in single cystathionine beta-synthase domain-containing protein from a bent to flat conformation upon adenosine monophosphate binding
J.Struct.Biol., 183, 2013
|
|
4EBR
 
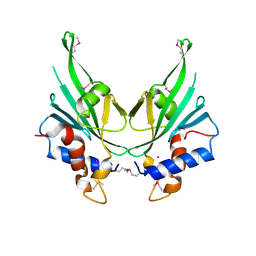 | | Crystal structure of Autophagic E2, Atg10 | | 分子名称: | MERCURY (II) ION, Ubiquitin-like-conjugating enzyme ATG10 | | 著者 | Hong, S.B, Kim, B.W, Kim, J.H, Song, H.K. | | 登録日 | 2012-03-24 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Structure of the autophagic E2 enzyme Atg10
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBA
 
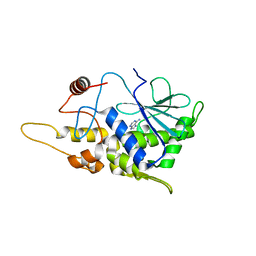 | | Structure of mutant RIP from barley seeds in complex with adenine | | 分子名称: | ADENINE, Protein synthesis inhibitor I | | 著者 | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | 登録日 | 2012-05-22 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4HNZ
 
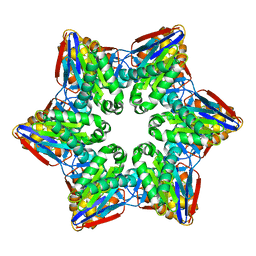 | |
4GQV
 
 | | Crystal structure of CBS-pair protein, CBSX1 from Arabidopsis thaliana | | 分子名称: | CBS domain-containing protein CBSX1, chloroplastic | | 著者 | Jeong, B.-C, Park, S.H, Yoo, K.S, Shin, J.S, Song, H.K. | | 登録日 | 2012-08-24 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.392 Å) | | 主引用文献 | Crystal structure of the single cystathionine beta-synthase domain-containing protein CBSX1 from Arabidopsis thaliana
Biochem.Biophys.Res.Commun., 430, 2013
|
|
4HO7
 
 | |
7Y6Z
 
 | | RLGSGG-AtPRT6 UBR box (I222) | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-06-21 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWG
 
 | | RSGSGG-AtPRT6 UBR box | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-05-26 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.832 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWE
 
 | | RRGSGG-AtPRT6 UBR box | | 分子名称: | E3 ubiquitin-protein ligase PRT6, MAGNESIUM ION, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-05-26 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWF
 
 | | RLGSGG-AtPRT6 UBR box (highest resolution) | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-05-26 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y70
 
 | | RLGSGG-AtPRT6 UBR box (P4332) | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-06-21 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWD
 
 | | Apo-AtPRT6 UBR box | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-05-26 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y6Y
 
 | | RLGSGG-AtPRT6 UBR box (C121) | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-06-21 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.543 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y6X
 
 | | RRGSGG-AtPRT6 UBR box (P32) | | 分子名称: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | 著者 | Kim, L, Song, H.K. | | 登録日 | 2022-06-21 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.196 Å) | | 主引用文献 | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7YRB
 
 | | UBR box of human UBR6 | | 分子名称: | F-box protein 11, isoform CRA_f, SULFATE ION, ... | | 著者 | Kim, B, Song, H.K. | | 登録日 | 2022-08-09 | | 公開日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Crystal structure of UBR box from human UBR6
To Be Published
|
|
7WG4
 
 | | DVAA-KlAte1 | | 分子名称: | Arginyltransferase, ZINC ION | | 著者 | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | 登録日 | 2021-12-28 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WG2
 
 | | EVAA-KlAte1 | | 分子名称: | Arginyltransferase, ZINC ION | | 著者 | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | 登録日 | 2021-12-28 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WFX
 
 | | EVAA-KlAte1 | | 分子名称: | Arginyltransferase, ZINC ION | | 著者 | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | 登録日 | 2021-12-27 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WG1
 
 | | DVAA-KlAte1 | | 分子名称: | Arginyltransferase, ZINC ION | | 著者 | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | 登録日 | 2021-12-27 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5HYY
 
 | |
7VGW
 
 | | Yeast gid10 with Pro-peptide | | 分子名称: | BJ4_G0041530.mRNA.1.CDS.1 | | 著者 | Shin, J.S, Park, S.H, Kim, L, Heo, J, Song, H.K. | | 登録日 | 2021-09-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of yeast Gid10 in complex with Pro/N-degron.
Biochem.Biophys.Res.Commun., 582, 2021
|
|
