4URP
 
 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | Descriptor: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | Authors: | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
3ZHD
 
 | |
4AEF
 
 | | THE CRYSTAL STRUCTURE OF THERMOSTABLE AMYLASE FROM THE PYROCOCCUS | | Descriptor: | NEOPULLULANASE (ALPHA-AMYLASE II) | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Yang, S.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2012-01-10 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A Novel Domain Arrangement in a Monomeric Cyclodextrin-Hydrolyzing Enzyme from the Hyperthermophile Pyrococcus Furiosus.
Biochim.Biophys.Acta, 1834, 2013
|
|
2VR5
 
 | | Crystal structure of Trex from Sulfolobus Solfataricus in complex with acarbose intermediate and glucose | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, GLYCEROL, GLYCOGEN OPERON PROTEIN GLGX, ... | | Authors: | Song, H.-N, Yoon, S.-M, Lee, S.-J, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
2VUY
 
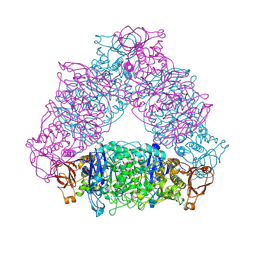 | | Crystal structure of Glycogen Debranching exzyme TreX from Sulfolobus solfatarius | | Descriptor: | GLYCOGEN OPERON PROTEIN GLGX | | Authors: | Song, H.-N, Yoon, S.-M, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
2VNC
 
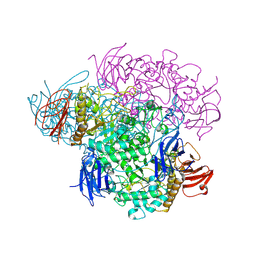 | | Crystal structure of Glycogen Debranching enzyme TreX from Sulfolobus solfataricus | | Descriptor: | GLYCOGEN OPERON PROTEIN GLGX | | Authors: | Song, H.-N, Yoon, S.-M, Cha, H, Park, K.-T, Woo, E.-J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
3ZHK
 
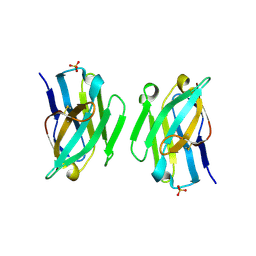 | |
3ZHL
 
 | |
2X6R
 
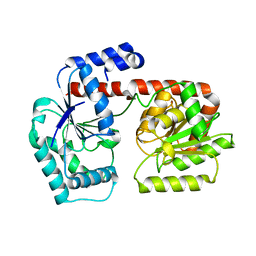 | | Crystal structure of trehalose synthase TreT from P.horikoshi produced by soaking in trehalose | | Descriptor: | TREHALOSE-SYNTHASE TRET | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lim, M.-Y, Lee, S.-B, Woo, E.-J. | | Deposit date: | 2010-02-19 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2WSK
 
 | | Crystal structure of Glycogen Debranching Enzyme GlgX from Escherichia coli K-12 | | Descriptor: | GLYCOGEN DEBRANCHING ENZYME, SULFATE ION | | Authors: | Song, H.-N, Park, J.-T, Jung, T.-Y, Park, K.-H, Woo, E.-J. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Rationale for the Short Branched Substrate Specificity of the Glycogen Debranching Enzyme Glgx.
Proteins, 78, 2010
|
|
2XMP
 
 | | Crystal structure of trehalose synthase TreT mutant E326A from P. horishiki in complex with UDP | | Descriptor: | TREHALOSE-SYNTHASE TRET, URIDINE-5'-DIPHOSPHATE | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2XA2
 
 | | Crystal structure of trehalose synthase TreT mutant E326A from P. horikoshii in complex with UDPG | | Descriptor: | TREHALOSE-SYNTHASE TRET, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2XA9
 
 | | Crystal structure of trehalose synthase TreT mutant E326A from P. horikoshii in complex with UDPG | | Descriptor: | TREHALOSE-SYNTHASE TRET, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2X6Q
 
 | | Crystal structure of trehalose synthase TreT from P.horikoshi | | Descriptor: | TREHALOSE-SYNTHASE TRET | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lim, M.-Y, Lee, S.-B, Woo, E.-J. | | Deposit date: | 2010-02-19 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
2XA1
 
 | | Crystal structure of trehalose synthase TreT from P.horikoshii (Seleno derivative) | | Descriptor: | TREHALOSE-SYNTHASE TRET | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-03-26 | | Release date: | 2010-10-13 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
4ATW
 
 | | The crystal structure of Arabinofuranosidase | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE DOMAIN PROTEIN | | Authors: | Dumbrepatil, A, Song, H.-N, Jung, T.-Y, Kim, T.-J, Woo, E.-J. | | Deposit date: | 2012-05-10 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Alpha-L-Arabinofuranosidase from Thermotoga Maritima Reveals Characteristics for Thermostability and Substrate Specificity.
J.Microbiol.Biotech., 22, 2012
|
|
4BBW
 
 | | The crystal structure of Sialidase VPI 5482 (BTSA) from Bacteroides thetaiotaomicron | | Descriptor: | SIALIDASE (NEURAMINIDASE) | | Authors: | Park, K.-H, Song, H.-N, Jung, T.-Y, Lee, M.-H, Woo, E.-J. | | Deposit date: | 2012-09-28 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Broad Substrate Specificity of Bacteroides Thetaiotaomicron Commensal Sialidase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4AP9
 
 | | Crystal structure of phosphoserine phosphatase from T. onnurineus in complex with NDSB-201 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Jung, T.-Y, Kim, Y.-S, Song, H.-N, Woo, E. | | Deposit date: | 2012-03-31 | | Release date: | 2012-12-26 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Identification of a Novel Ligand Binding Site in Phosphoserine Phosphatase from the Hyperthermophilic Archaeon Thermococcus Onnurineus.
Proteins, 81, 2013
|
|
4B6J
 
 | |
2X1I
 
 | | glycoside hydrolase family 77 4-alpha-glucanotransferase from thermus brockianus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 4-ALPHA-GLUCANOTRANSFERASE, PHOSPHATE ION, ... | | Authors: | Yoon, S.-M, Jung, J.-H, Jung, T.-Y, Song, H.-N, Park, C.-S, Woo, E.-J. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and Functional Analysis of Substrate Recognition by the 250S Loop in Amylomaltase from Thermus Brockianus.
Proteins, 79, 2011
|
|
2WC7
 
 | | Crystal structure of Nostoc Punctiforme Debranching Enzyme(NPDE)(Acarbose soaked) | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Dumbrepatil, A.-B, Song, H.-N, Choi, J.-H, Park, K.-H, Woo, E.-J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Features of the Nostoc Punctiforme Debranching Enzyme Reveal the Basis of its Mechanism and Substrate Specificity.
Proteins, 78, 2010
|
|
