7C4W
 
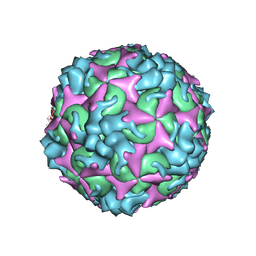 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3S2Y
 
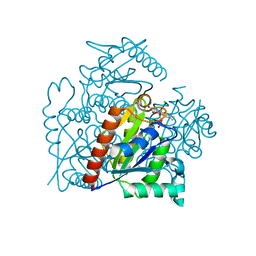 | | Crystal structure of a chromate/uranium reductase from Gluconacetobacter hansenii | | Descriptor: | CHLORIDE ION, Chromate reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jin, H, Zhang, Y, Buchko, G.W, Li, P, Squier, T.C, Robinson, H, Varnum, S.M, Long, P.E. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Structure Determination and Functional Analysis of a Chromate Reductase from Gluconacetobacter hansenii.
Plos One, 7, 2012
|
|
2LT7
 
 | | Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B3G
 
 | | crystal structure of Ighmbp2 helicase in complex with RNA | | Descriptor: | DNA-BINDING PROTEIN SMUBP-2, RNA (5'-(AP*AP*AP*AP*AP*AP*AP*AP*AP)-3') | | Authors: | Lim, S.C, Song, H. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Ighmbp2 Helicase Structure Reveals the Molecular Basis for Disease-Causing Mutations in Dmsa1.
Nucleic Acids Res., 40, 2012
|
|
5EXS
 
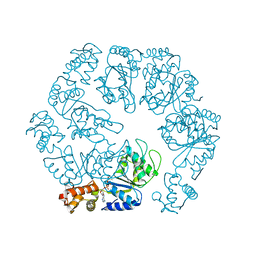 | |
5EXX
 
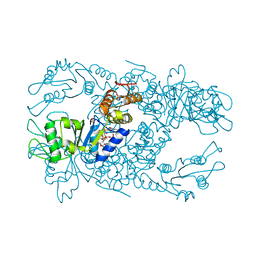 | | AAA+ ATPase FleQ from Pseudomonas aeruginosa bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Transcriptional regulator FleQ | | Authors: | Navarro, M.V.A.S, Sondermann, H, Krasteva, P.V. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.311 Å) | | Cite: | Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3HVA
 
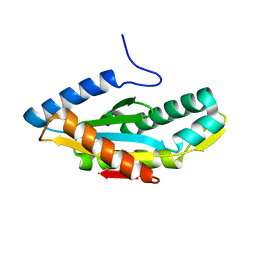 | |
1AUR
 
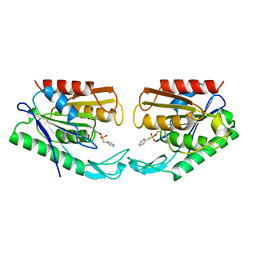 | |
4OZW
 
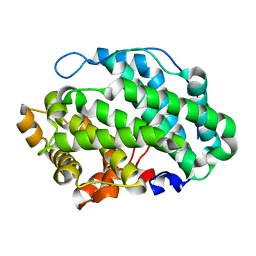 | |
4FGP
 
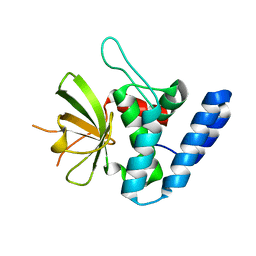 | | Legionella pneumophila LapG (EGTA-treated) | | Descriptor: | Periplasmic protein | | Authors: | Chatterjee, D, Boyd, C.D, O'Toole, G.A, Sondermann, H. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural characterization of a conserved, calcium-dependent periplasmic protease from Legionella pneumophila.
J.Bacteriol., 194, 2012
|
|
2HTS
 
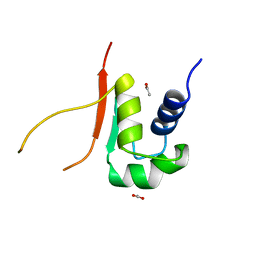 | |
1O55
 
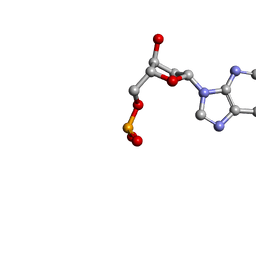 | | MOLECULAR STRUCTURE OF TWO CRYSTAL FORMS OF CYCLIC TRIADENYLIC ACID AT 1 ANGSTROM RESOLUTION | | Descriptor: | COBALT (II) ION, DNA (5'-CD(*AP*AP*AP)-3') | | Authors: | Gao, Y.G, Robinson, H, Guan, Y, Liaw, Y.C, van Boom, J.H, van der Marel, G.A, Wang, A.H. | | Deposit date: | 2003-08-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Molecular structure of two crystal forms of cyclic triadenylic acid at 1A resolution.
J.Biomol.Struct.Dyn., 16, 1998
|
|
1UCS
 
 | | Type III Antifreeze Protein RD1 from an Antarctic Eel Pout | | Descriptor: | Antifreeze peptide RD1 | | Authors: | Ko, T.-P, Robinson, H, Gao, Y.-G, Cheng, C.-H.C, DeVries, A.L, Wang, A.H.-J. | | Deposit date: | 2003-04-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.62 Å) | | Cite: | The refined crystal structure of an eel pout type III antifreeze protein RD1 at 0.62-A resolution reveals structural microheterogeneity of protein and solvation.
Biophys.J., 84, 2003
|
|
1AUO
 
 | | CARBOXYLESTERASE FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | CARBOXYLESTERASE | | Authors: | Kim, K.K, Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of carboxylesterase from Pseudomonas fluorescens, an alpha/beta hydrolase with broad substrate specificity.
Structure, 5, 1997
|
|
2HD1
 
 | | Crystal structure of PDE9 in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, Phosphodiesterase 9A, ... | | Authors: | Huai, Q, Wang, H, Zhang, W, Colman, R.W, Robinson, H, Ke, H. | | Deposit date: | 2006-06-19 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of phosphodiesterase 9 shows orientation variation of inhibitor IBMX binding
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5FR3
 
 | | X-ray crystal structure of aggregation-resistant protective antigen of Bacillus anthracis (mutant S559L T576E) | | Descriptor: | CALCIUM ION, GLYCEROL, PROTECTIVE ANTIGEN | | Authors: | Ganesan, A, Siekierska, A, Beerten, J, Brams, M, van Durme, J, De Baets, G, van der Kant, R, Gallardo, R, Ramakers, M, Langenberg, T, Wilkinson, H, De Smet, F, Ulens, C, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural Hot Spots for the Solubility of Globular Proteins
Nat.Commun., 7, 2016
|
|
4P7Q
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1HXY
 
 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCAL ENTEROTOXIN H IN COMPLEX WITH HUMAN MHC CLASS II | | Descriptor: | ENTEROTOXIN H, HEMAGGLUTININ, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Petersson, K, Hakansson, M, Nilsson, H, Forsberg, G, Svensson, L.A, Liljas, A, Walse, B. | | Deposit date: | 2001-01-17 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Superantigen Bound to MHC Class II Displays Zinc and Peptide Dependence
Embo J., 20, 2001
|
|
3CSX
 
 | |
1Q2U
 
 | | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Huai, Q, Sun, Y, Wang, H, Chin, L.S, Li, L, Robinson, H, Ke, H. | | Deposit date: | 2003-07-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease
Febs Lett., 549, 2003
|
|
5EXT
 
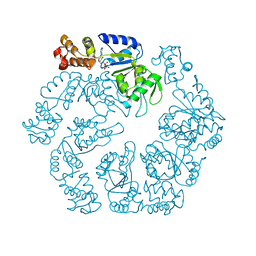 | |
1RGO
 
 | | Structural Basis for Recognition of the mRNA Class II AU-Rich Element by the Tandem Zinc Finger Domain of TIS11d | | Descriptor: | Butyrate response factor 2, RNA (5'-R(*UP*UP*AP*UP*UP*UP*AP*UP*U)-3'), ZINC ION | | Authors: | Hudson, B.P, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-11-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d.
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1C78
 
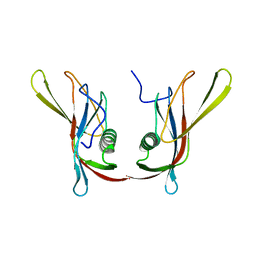 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C77
 
 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C79
 
 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
