8HRP
 
 | |
8HRR
 
 | |
8HRT
 
 | |
8HRQ
 
 | |
8HRS
 
 | |
8HRO
 
 | |
8JB1
 
 | |
6IUM
 
 | |
6IUN
 
 | |
1I3W
 
 | | ACTINOMYCIN D BINDING TO CGATCGATCG | | Descriptor: | 5'-D(*C*GP*AP*TP*CP*GP*AP*(BRU)P*CP*GP)-3', ACTINOMYCIN D | | Authors: | Robinson, H, Gao, Y.-G, Yang, X.-L, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 2001-02-17 | | Release date: | 2001-05-21 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Analysis of a Novel Complex of Actinomycin D Bound to the DNA Decamer Cgatcgatcg.
Biochemistry, 40, 2001
|
|
7NEF
 
 | | Fucosylated linear peptide Fln65 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fln65, ... | | Authors: | Personne, H, Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A mixed chirality alpha-helix in a stapled bicyclic and a linear antimicrobial peptide revealed by X-ray crystallography.
Rsc Chem Biol, 2, 2021
|
|
7NEW
 
 | | Fucosylated heterochiral linear peptide Fdln69 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 2.0 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Personne, H, Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A mixed chirality alpha-helix in a stapled bicyclic and a linear antimicrobial peptide revealed by X-ray crystallography.
Rsc Chem Biol, 2, 2021
|
|
6QZY
 
 | | full length OphA V406P in complex with SAH | | Descriptor: | ASN-GLY-PHE-PRO-TRP-MVA-ILE-MVA-VAL-GLY-PRO-ILE-GLY, MAGNESIUM ION, Peptide N-methyltransferase, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6QZZ
 
 | | full length OphA V404E in complex with SAH | | Descriptor: | Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6R00
 
 | | OphA DeltaC6 V404F complex with SAH | | Descriptor: | PHE-PRO-TRP-MVA-ILE-MVA-PHE-GLY-VAL-ILE-GLY-VAL-ILE-GLY, Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
5YN3
 
 | |
5X5T
 
 | |
5X7N
 
 | |
5X5U
 
 | |
5IJZ
 
 | |
5X7M
 
 | |
1HSE
 
 | | H253M N TERMINAL LOBE OF HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Nicholson, H, Anderson, B.F, Baker, E.N. | | Deposit date: | 1996-12-11 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis of the histidine ligand in human lactoferrin: iron binding properties and crystal structure of the histidine-253-->methionine mutant.
Biochemistry, 36, 1997
|
|
5HXX
 
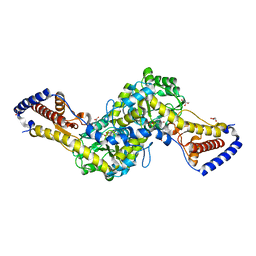 | | Crystal structure of AspAT from Corynebacterium glutamicum | | Descriptor: | 2-OXOGLUTARIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, ... | | Authors: | Son, H.-F, Kim, K.-J. | | Deposit date: | 2016-01-31 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into a novel class of aspartate aminotransferase from Corynebacterium glutamicum
To be published
|
|
5E66
 
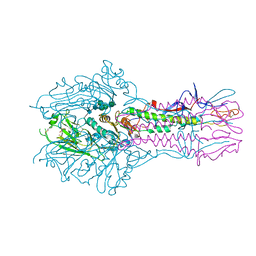 | | The complex structure of Hemagglutinin-esterase-fusion mutant protein from the influenza D virus with receptor analog 9-N-Ac-Sia | | Descriptor: | (6R)-5-acetamido-6-[(1S,2S)-3-acetamido-1,2-dihydroxypropyl]-3,5-dideoxy-beta-L-threo-hex-2-ulopyranosonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5IWQ
 
 | |
