4UX7
 
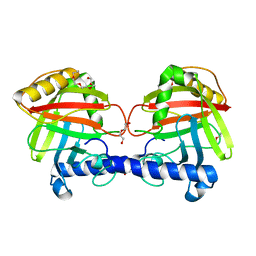 | | Structure of a Clostridium difficile sortase | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PUTATIVE PEPTIDASE C60B, ... | | Authors: | Chambers, C.J, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2014-08-19 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Function of a Clostridium Difficile Sortase Enzyme.
Sci.Rep., 5, 2015
|
|
2BOV
 
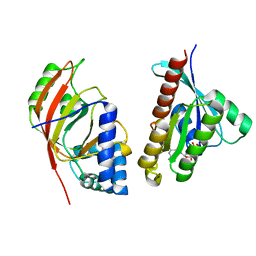 | | Molecular recognition of an ADP-ribosylating Clostridium botulinum C3 exoenzyme by RalA GTPase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Holbourn, K.P, Sutton, J.M, Evans, H.R, Shone, C.C, Acharya, K.R. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Molecular Recognition of an Adp-Ribosylating Clostridium Botulinum C3 Exoenzyme by Rala Gtpase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1UZI
 
 | | C3 EXOENZYME FROM CLOSTRIDIUM BOTULINUM, TETRAGONAL FORM | | Descriptor: | CYCLO-TETRAMETAVANADATE, GLYCEROL, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Evans, H.R, Holloway, D.E, Sutton, J.M, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | C3 Exoenzyme from Clostridium Botulinum: Structure of a Tetragonal Crystal Form and a Reassessment of Nad-Induced Flexure
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OJZ
 
 | | The crystal structure of C3stau2 from S. aureus with NAD | | Descriptor: | ADP-RIBOSYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-16 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
5NJL
 
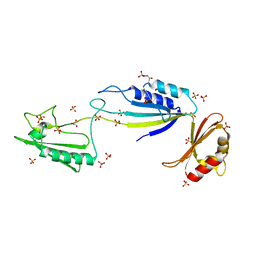 | | Cwp2 from Clostridium difficile | | Descriptor: | Cell surface protein (Putative S-layer protein), SULFATE ION | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-03-29 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cwp2 from Clostridium difficile exhibits an extended three domain fold and cell adhesion in vitro.
FEBS J., 284, 2017
|
|
5OQ3
 
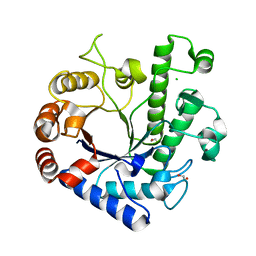 | | High resolution structure of the functional region of Cwp19 from Clostridium difficile | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cwp19, ... | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile.
FEBS J., 284, 2017
|
|
5OQ2
 
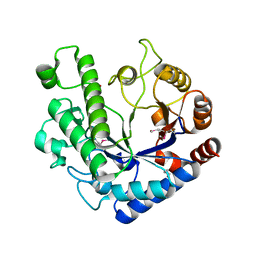 | | Se-SAD structure of the functional region of Cwp19 from Clostridium difficile | | Descriptor: | 1,2-ETHANEDIOL, Cwp19, PHOSPHATE ION | | Authors: | Bradshaw, W.J, Kirby, J.M, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile.
FEBS J., 284, 2017
|
|
1OJQ
 
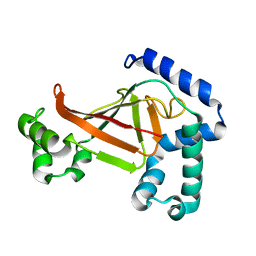 | | The crystal structure of C3stau2 from S. aureus | | Descriptor: | ADP-RIBOSYLTRANSFERASE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-15 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
4D5A
 
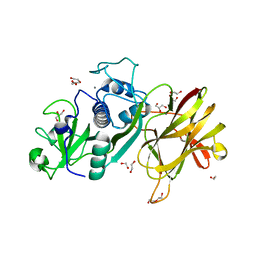 | | Clostridial Cysteine protease Cwp84 C116A after propeptide cleavage | | Descriptor: | CALCIUM ION, CELL SURFACE PROTEIN (PUTATIVE CELL SURFACE-ASSOCIATED CYSTEINE PROTEASE), GLYCEROL, ... | | Authors: | Bradshaw, W.J, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2014-11-03 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cwp84, a Clostridium Difficile Cysteine Protease, Exhibits Conformational Flexibility in the Absence of its Propeptide
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4D59
 
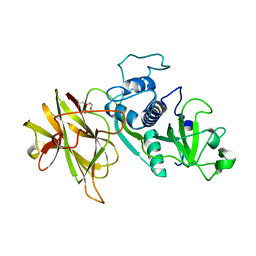 | | Clostridial Cysteine protease Cwp84 C116A after propeptide cleavage | | Descriptor: | CALCIUM ION, CELL SURFACE PROTEIN (PUTATIVE CELL SURFACE-ASSOCIATED CYSTEINE PROTEASE), O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Bradshaw, W.J, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2014-11-03 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cwp84, a Clostridium Difficile Cysteine Protease, Exhibits Conformational Flexibility in the Absence of its Propeptide
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4CI7
 
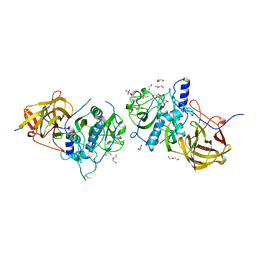 | | The crystal structure of the cysteine protease and lectin-like domains of Cwp84, a surface layer associated protein of Clostridium difficile | | Descriptor: | CALCIUM ION, CELL SURFACE PROTEIN (PUTATIVE CELL SURFACE-ASSOCIATED CYSTEINE PROTEASE), GLYCEROL, ... | | Authors: | Bradshaw, W.J, Kirby, J.M, Thiyagarajan, N, Chambers, C.J, Davies, A.H, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structure of the Cysteine Protease and Lectin-Like Domains of Cwp84, a Surface Layer-Associated Protein from Clostridium Difficile
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2WN5
 
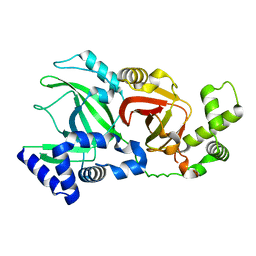 | |
2WN7
 
 | | Structural Basis for Substrate Recognition in the Enzymatic Component of ADP-ribosyltransferase Toxin CDTa from Clostridium difficile | | Descriptor: | ADP-RIBOSYLTRANSFERASE ENZYMATIC COMPONENT, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sundriyal, A, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2009-07-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Substrate Recognition in the Enzymatic Component of Adp-Ribosyltransferase Toxin Cdta from Clostridium Difficile.
J.Biol.Chem., 284, 2009
|
|
2WN8
 
 | |
2WN4
 
 | |
2WN6
 
 | | Structural Basis for Substrate Recognition in the Enzymatic Component of ADP-ribosyltransferase Toxin CDTa from Clostridium difficile | | Descriptor: | ADP-RIBOSYLTRANSFERASE ENZYMATIC COMPONENT, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundriyal, A, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2009-07-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for Substrate Recognition in the Enzymatic Component of Adp-Ribosyltransferase Toxin Cdta from Clostridium Difficile.
J.Biol.Chem., 284, 2009
|
|
