8I34
 
 | | The crystal structure of EPD-BCP1 from a marine sponge | | Descriptor: | (2~{Z},4~{E},6~{E},8~{E},10~{E},12~{E},14~{E},16~{E})-4,8,13,17-tetramethyl-3-oxidanyl-19-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]-1-[(1~{R},4~{S})-1,2,2-trimethyl-4-oxidanyl-cyclopentyl]nonadeca-2,4,6,8,10,12,14,16-octaen-18-yn-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASTAXANTHIN, ... | | Authors: | Shomura, Y, Kawasaki, S. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | An ependymin-related blue carotenoprotein decorates marine blue sponge.
J.Biol.Chem., 299, 2023
|
|
4P7X
 
 | | L-pipecolic acid-bound L-proline cis-4-hydroxylase | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-OXOGLUTARIC ACID, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | Authors: | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | Deposit date: | 2014-03-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
1Q3S
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (FormIII crystal complexed with ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
4P7W
 
 | | L-proline-bound L-proline cis-4-hydroxylase | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, L-proline cis-4-hydroxylase, ... | | Authors: | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | Deposit date: | 2014-03-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
1XQS
 
 | | Crystal structure of the HspBP1 core domain complexed with the fragment of Hsp70 ATPase domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, HSPBP1 protein, Heat shock 70 kDa protein 1 | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
1XQR
 
 | | Crystal structure of the HspBP1 core domain | | Descriptor: | HspBP1 protein | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
1Q2V
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form) | | Descriptor: | SULFATE ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Yohda, M, Maruyama, T, Miki, K. | | Deposit date: | 2003-07-26 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Q3R
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) | | Descriptor: | SULFATE ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Q3Q
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (two-point mutant complexed with AMP-PNP) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Z1F
 
 | | Crystal structure of stilbene synthase from Arachis hypogaea (resveratrol-bound form) | | Descriptor: | CITRIC ACID, RESVERATROL, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
1Z1E
 
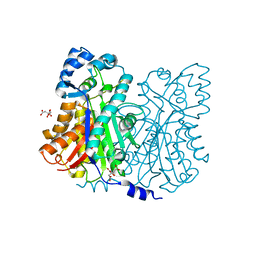 | | Crystal structure of stilbene synthase from Arachis hypogaea | | Descriptor: | CITRIC ACID, stilbene synthase | | Authors: | Shomura, Y, Torayama, I, Suh, D.Y, Xiang, T, Kita, A, Sankawa, U, Miki, K. | | Deposit date: | 2005-03-03 | | Release date: | 2005-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of stilbene synthase from Arachis hypogaea
Proteins, 60, 2005
|
|
2Z1T
 
 | |
2Z1U
 
 | |
3VTI
 
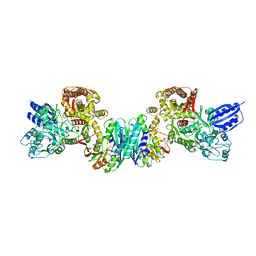 | | Crystal structure of HypE-HypF complex | | Descriptor: | FE (III) ION, Hydrogenase maturation factor, MAGNESIUM ION, ... | | Authors: | Shomura, Y, Higuchi, Y. | | Deposit date: | 2012-05-30 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for the reaction mechanism of S-carbamoylation of HypE by HypF in the maturation of [NiFe]-hydrogenases
J.Biol.Chem., 287, 2012
|
|
3AYX
 
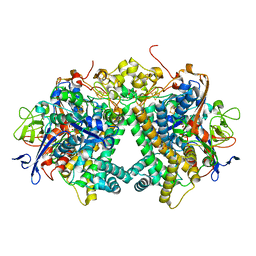 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an H2-reduced condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
3AYZ
 
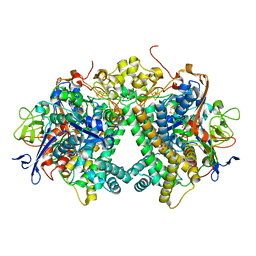 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an air-oxidized condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
5Y34
 
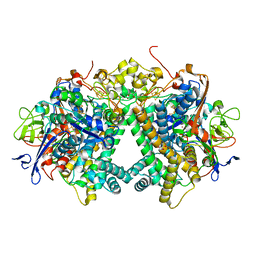 | | Membrane-bound respiratory [NiFe]-hydrogenase from Hydrogenovibrio marinus in a ferricyanide-oxidized condition | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2017-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
3VTH
 
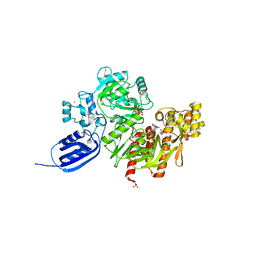 | |
3VMM
 
 | |
5XF9
 
 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
5XFA
 
 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the H2-reduced state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
3KLJ
 
 | | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-dependent dehydrogenase, NirB-family (N-terminal domain) | | Authors: | Nishikawa, K, Shomura, Y, Kawasaki, S, Niimura, Y, Higuchi, Y. | | Deposit date: | 2009-11-08 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum: a key component of the dioxygen scavenging system in obligatory anaerobes.
Proteins, 78, 2010
|
|
5AWI
 
 | | Domain-swapped cytochrome cb562 dimer | | Descriptor: | HEME C, SULFATE ION, Soluble cytochrome b562, ... | | Authors: | Miyamoto, T, Kuribayashi, M, Nagao, S, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Domain-swapped cytochrome cb562 dimer and its nanocage encapsulating a Zn-SO4 cluster in the internal cavity
Chem Sci, 2015
|
|
5AUR
 
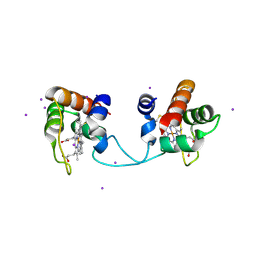 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at N-terminal region | | Descriptor: | Cytochrome c-552, HEME C, IODIDE ION | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Kamikubo, H, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
5AUS
 
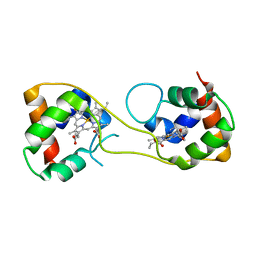 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at C-terminal region | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
