1TET
 
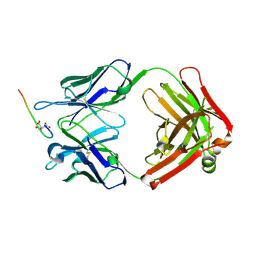 | |
1CN1
 
 | | CRYSTAL STRUCTURE OF DEMETALLIZED CONCANAVALIN A. THE METAL-BINDING REGION | | Descriptor: | CONCANAVALIN A | | Authors: | Shoham, M, Yonath, A, Sussman, J.L, Moult, J, Traub, W, Gilboa(Kalb), A.J. | | Deposit date: | 1981-12-21 | | Release date: | 1982-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of demetallized concanavalin A: the metal-binding region.
J.Mol.Biol., 131, 1979
|
|
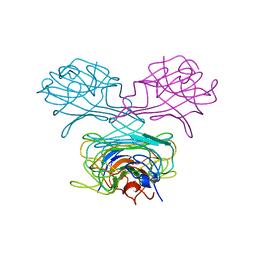
2GL2
 
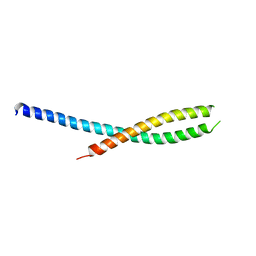 | | Crystal structure of the tetra mutant (T66G,R67G,F68G,Y69G) of bacterial adhesin FadA | | Descriptor: | adhesion A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2006-04-04 | | Release date: | 2007-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary X-ray data of the FadA adhesin from Fusobacterium nucleatum.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1A3Z
 
 | | REDUCED RUSTICYANIN AT 1.9 ANGSTROMS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Zhao, D, Shoham, M. | | Deposit date: | 1998-01-27 | | Release date: | 1998-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rusticyanin: Extremes in acid stability and redox potential explained by the crystal structure.
Biophys.J., 74, 1998
|
|
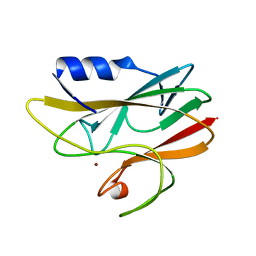
1BXZ
 
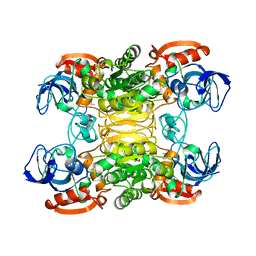 | | CRYSTAL STRUCTURE OF A THERMOPHILIC ALCOHOL DEHYDROGENASE SUBSTRATE COMPLEX FROM THERMOANAEROBACTER BROCKII | | Descriptor: | 2-BUTANOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, C, Heatwole, J, Soelaiman, S, Shoham, M. | | Deposit date: | 1998-10-09 | | Release date: | 2000-02-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of a thermophilic alcohol dehydrogenase substrate complex suggests determinants of substrate specificity and thermostability.
Proteins, 37, 1999
|
|
1DOI
 
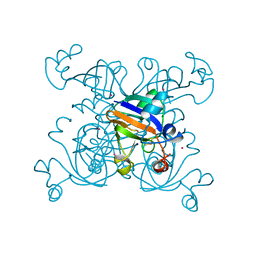 | | 2FE-2S FERREDOXIN FROM HALOARCULA MARISMORTUI | | Descriptor: | 2FE-2S FERREDOXIN, FE2/S2 (INORGANIC) CLUSTER, POTASSIUM ION | | Authors: | Frolow, F, Harel, M, Sussman, J.L, Shoham, M. | | Deposit date: | 1996-04-08 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into protein adaptation to a saturated salt environment from the crystal structure of a halophilic 2Fe-2S ferredoxin.
Nat.Struct.Biol., 3, 1996
|
|
3ETW
 
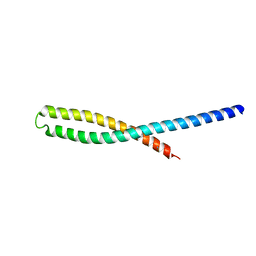 | | Crystal Structure of bacterial adhesin FadA | | Descriptor: | Adhesin A, THIOCYANATE ION | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETY
 
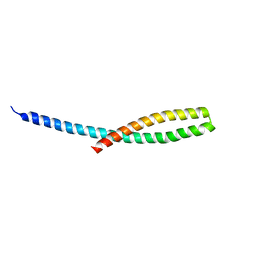 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
1RCY
 
 | | RUSTICYANIN (RC) FROM THIOBACILLUS FERROOXIDANS | | Descriptor: | COPPER (II) ION, RUSTICYANIN | | Authors: | Walter, R.L, Friedman, A.M, Ealick, S.E, Blake II, R.C, Proctor, P, Shoham, M. | | Deposit date: | 1996-04-10 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability.
J.Mol.Biol., 263, 1996
|
|
3ETX
 
 | | Crystal structure of bacterial adhesin FadA L14A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
3ETZ
 
 | | Crystal structure of bacterial adhesin FadA L76A mutant | | Descriptor: | Adhesin A | | Authors: | Nithianantham, S, Xu, M, Wu, N, Shoham, M, Han, Y.W. | | Deposit date: | 2008-10-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
J.Biol.Chem., 284, 2009
|
|
1YTV
 
 | | Maltose-binding protein fusion to a C-terminal fragment of the V1a vasopressin receptor | | Descriptor: | Maltose-binding periplasmic protein, Vasopressin V1a receptor, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Adikesavan, N.V, Mahmood, S.S, Stanley, S, Xu, Z, Wu, N, Thibonnier, M, Shoham, M. | | Deposit date: | 2005-02-11 | | Release date: | 2005-04-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-terminal segment of the V1R vasopressin receptor is unstructured in the crystal structure of its chimera with the maltose-binding protein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2B5U
 
 | | Crystal Structure Of Colicin E3 V206C Mutant In Complex With Its Immunity Protein | | Descriptor: | CITRIC ACID, Colicin E3, Colicin E3 immunity protein | | Authors: | Nallini Vijayarangan, A, Nithianantham, S, Nan, W, Jakes, K, Shoham, M. | | Deposit date: | 2005-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Of Colicin E3 In Complex With Its Immunity Protein
TO BE PUBLISHED
|
|
3EIP
 
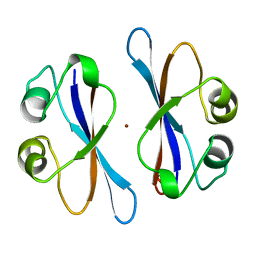 | | CRYSTAL STRUCTURE OF COLICIN E3 IMMUNITY PROTEIN: AN INHIBITOR TO A RIBOSOME-INACTIVATING RNASE | | Descriptor: | PROTEIN (COLICIN E3 IMMUNITY PROTEIN), ZINC ION | | Authors: | Li, C, Zhao, D, Djebli, A, Shoham, M. | | Deposit date: | 1999-03-29 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase.
Structure Fold.Des., 7, 1999
|
|
1M11
 
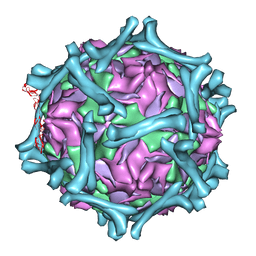 | | structural model of human decay-accelerating factor bound to echovirus 7 from cryo-electron microscopy | | Descriptor: | COAT PROTEIN VP1, COAT PROTEIN VP2, COAT PROTEIN VP3, ... | | Authors: | He, Y, Lin, F, Chipman, P.R, Bator, C.M, Baker, T.S, Shoham, M, Kuhn, R.J, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2002-06-17 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JCH
 
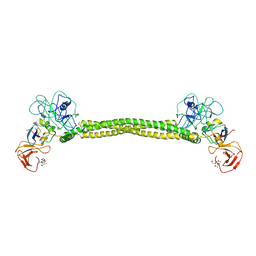 | | Crystal Structure of Colicin E3 in Complex with its Immunity Protein | | Descriptor: | CITRIC ACID, COLICIN E3, COLICIN E3 IMMUNITY PROTEIN, ... | | Authors: | Soelaiman, S, Jakes, K, Wu, N, Li, C, Shoham, M. | | Deposit date: | 2001-06-09 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of colicin E3: implications for cell entry and ribosome inactivation.
Mol.Cell, 8, 2001
|
|
