2DK8
 
 | | Solution structure of rpc34 subunit in RNA polymerase III from mouse | | Descriptor: | DNA-directed RNA polymerase III 39 kDa polypeptide | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of rpc34 subunit in RNA polymerase III from mouse
To be Published
|
|
2DNO
 
 | | Solution structure of RNA binding domain in Trinucleotide repeat containing 4 variant | | Descriptor: | trinucleotide repeat containing 4 variant | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in Trinucleotide repeat containing 4 variant
To be Published
|
|
2DNH
 
 | | Solution structure of RNA binding domain in Bruno-like 5 RNA binding protein | | Descriptor: | Bruno-like 5, RNA binding protein | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in Bruno-like 5 RNA binding protein
To be Published
|
|
2D9P
 
 | | Solution structure of RNA binding domain 4 in Polyadenylation binding protein 3 | | Descriptor: | Polyadenylate-binding protein 3 | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-12 | | Release date: | 2006-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain 4 in Polyadenylation binding
protein 3
To be Published
|
|
2D9N
 
 | | Solution structure of CCCH type zinc-finger domain 2 in Cleavage and polyadenylation specificity factor | | Descriptor: | Cleavage and polyadenylation specificity factor, 30 kDa subunit, ZINC ION | | Authors: | Abe, C, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-12 | | Release date: | 2006-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of CCCH type zinc-finger domain 2 in
Cleavage and polyadenylation specificity factor
To be Published
|
|
2D9M
 
 | | Solution structure of CCCH type zinc-finger domain 3 in zinc finger CCCH-type domain containing 7A | | Descriptor: | ZINC ION, Zinc finger CCCH-type domain containing protein 7A | | Authors: | Abe, C, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-12 | | Release date: | 2006-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of CCCH type zinc-finger domain 3 in zinc finger CCCH-type domain containing 7A
To be Published
|
|
2DB4
 
 | | Crystal structure of rotor ring with DCCD of the V- ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DICYCLOHEXYLUREA, SODIUM ION, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2DUL
 
 | | Crystal structure of tRNA G26 methyltransferase Trm1 in apo form from Pyrococcus horikoshii | | Descriptor: | GLYCEROL, N(2),N(2)-dimethylguanosine tRNA methyltransferase | | Authors: | Ihsanawati, Shirouzu, M, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-24 | | Release date: | 2007-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of tRNA N(2),N(2)-Guanosine Dimethyltransferase Trm1 from Pyrococcus horikoshii
J.Mol.Biol., 383, 2008
|
|
2EJT
 
 | | Complex structure of Trm1 from Pyrococcus horikoshii with S-adenosyl-L-Methionine | | Descriptor: | GLYCEROL, N(2),N(2)-dimethylguanosine tRNA methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Ihsanawati, Shirouzu, M, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of tRNA N(2),N(2)-Guanosine Dimethyltransferase Trm1 from Pyrococcus horikoshii
J.Mol.Biol., 383, 2008
|
|
2EJU
 
 | | Complex structure of Trm1 from Pyrococcus horikoshii with S-adenosyl-L-Homocystein | | Descriptor: | GLYCEROL, N(2),N(2)-dimethylguanosine tRNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ihsanawati, Shirouzu, M, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of tRNA N(2),N(2)-Guanosine Dimethyltransferase Trm1 from Pyrococcus horikoshii
J.Mol.Biol., 383, 2008
|
|
2DST
 
 | | Crystal Structure Analysis of TT1977 | | Descriptor: | Hypothetical protein TTHA1544 | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Shirouzu, M, Chen, L, Liu, Z.J, Wang, B.C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-06 | | Release date: | 2007-01-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the minimized alpha/beta-hydrolase fold protein from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1UEB
 
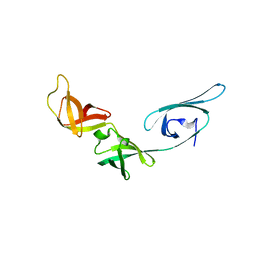 | | Crystal structure of translation elongation factor P from Thermus thermophilus HB8 | | Descriptor: | elongation factor P | | Authors: | Hanawa-Suetsugu, K, Sekine, S, Sakai, H, Hori-Takemoto, C, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-09 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of elongation factor P from Thermus thermophilus HB8
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1UFR
 
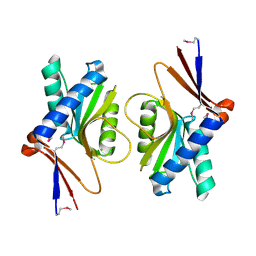 | | Crystal Structure of TT1027 from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, pyr mRNA-binding attenuation protein | | Authors: | Matsuura, T, Sakai, H, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-08 | | Release date: | 2003-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of TT1027 from Thermus thermophilus HB8
To be Published
|
|
1UFL
 
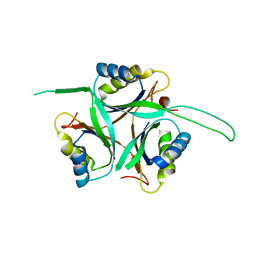 | | Crystal Structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wang, H, Sakai, H, Hori-Takemoto, C, Kaminishi, T, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-31 | | Release date: | 2003-11-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8.
J.Struct.Biol., 149, 2005
|
|
1VFJ
 
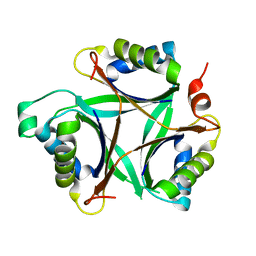 | | Crystal structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | nitrogen regulatory protein p-II | | Authors: | Wang, H, Sakai, H, Takemoto-Hori, C, Kaminishi, T, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8
J.STRUCT.BIOL., 149, 2005
|
|
1WN9
 
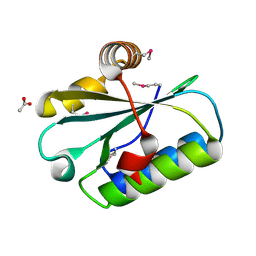 | | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8 | | Descriptor: | ACETIC ACID, the hypothetical protein (TT1805) | | Authors: | Pioszak, A.A, Kishishita, S, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-28 | | Release date: | 2005-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8
To be Published
|
|
1WDE
 
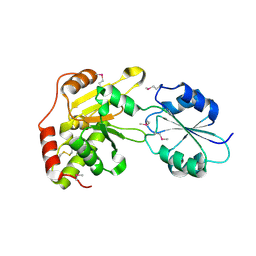 | | Crystal structure of the conserved hypothetical protein APE0931 from Aeropyrum pernix K1 | | Descriptor: | Probable diphthine synthase | | Authors: | Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-13 | | Release date: | 2004-11-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two archaeal diphthine synthases: insights into the post-translational modification of elongation factor 2.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1WXX
 
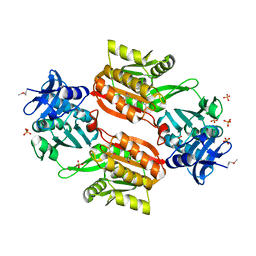 | | Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, hypothetical protein TTHA1280 | | Authors: | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-02 | | Release date: | 2005-08-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1WNA
 
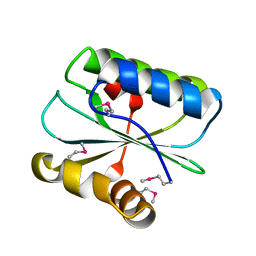 | | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8 | | Descriptor: | the hypothetical protein (TT1805) | | Authors: | Pioszak, A.A, Kishishita, S, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-28 | | Release date: | 2005-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the hypothetical protein TT1805 from Thermus thermophillus HB8
To be Published
|
|
1WWS
 
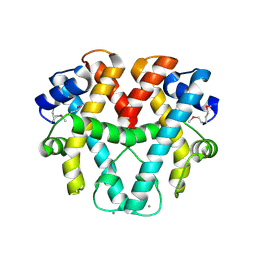 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | CALCIUM ION, hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
to be published
|
|
1WWI
 
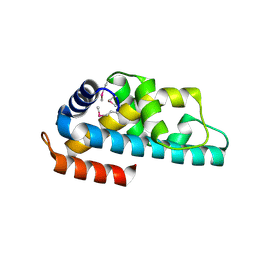 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
1WD7
 
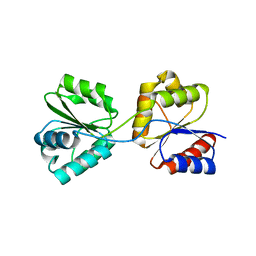 | | Crystal Structure of Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (Wild type, Native, Form-2 crystal) | | Descriptor: | Uroporphyrinogen III Synthase | | Authors: | Mizohata, E, Matsuura, T, Sakai, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-12 | | Release date: | 2004-11-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Uroporphyrinogen III Synthase from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 (Wild type, Native, Form-2 crystal)
to be published
|
|
1WXW
 
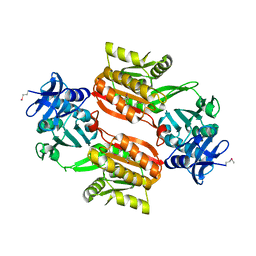 | | Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8 | | Descriptor: | HEXANE-1,6-DIOL, hypothetical protein TTHA1280 | | Authors: | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-02 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1WDZ
 
 | | Crystal structure of RCB domain of IRSp53 | | Descriptor: | insulin receptor substrate p53 | | Authors: | Murayama, K, Suetsugu, S, Seto, A, Shirouzu, M, Takenawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-21 | | Release date: | 2005-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of RCB domain of IRSp53
TO BE PUBLISHED
|
|
1WG8
 
 | |
