1CVM
 
 | | CADMIUM INHIBITED CRYSTAL STRUCTURE OF PHYTASE FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | CADMIUM ION, CALCIUM ION, PHYTASE | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-24 | | Release date: | 2000-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
1GS3
 
 | |
1H6L
 
 | |
1QLG
 
 | | Crystal structure of phytase with magnesium from Bacillus amyloliquefaciens | | Descriptor: | 3-PHYTASE, CALCIUM ION, MAGNESIUM ION | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-31 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of a Novel, Thermostable Phytase in Partially and Fully Calcium-Loaded States
Nat.Struct.Biol., 7, 2000
|
|
1O9N
 
 | |
1OBI
 
 | |
1OBK
 
 | |
1OCM
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COVALENTLY COMPLEXED WITH PYROPHOSPHATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, PYROPHOSPHATE 2- | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1O9O
 
 | |
1O9Q
 
 | |
1OCK
 
 | |
1O9P
 
 | |
1OCL
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COMPLEXED WITH MALONATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, MALONIC ACID | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1OBL
 
 | |
1OBJ
 
 | |
1OCH
 
 | |
3AJV
 
 | | Splicing endonuclease from Aeropyrum pernix | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative uncharacterized protein, ... | | Authors: | Yoshinari, S, Watanabe, Y, Okuda, M, Shiba, T, Inaoka, K.D, Kurisu, G. | | Deposit date: | 2010-06-19 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Conserved Lysine Residue in the Crenarchaea-Specific Loop is Important for the Crenarchaeal Splicing Endonuclease Activity.
J.Mol.Biol., 405, 2011
|
|
2DEP
 
 | |
2DRR
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263N mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRS
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263S mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRO
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263C mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2DRQ
 
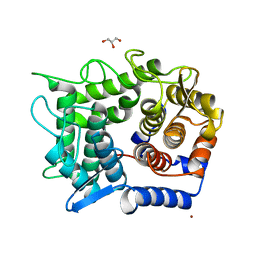 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase D263G mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2006-06-12 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 2009
|
|
2RV9
 
 | |
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
2RVA
 
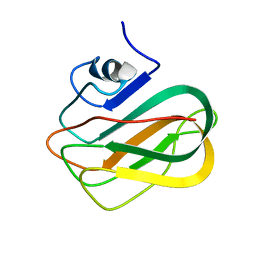 | |
