5YRT
 
 | |
5YSN
 
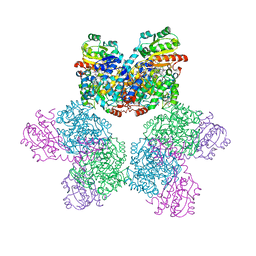 | | Ethanolamine ammonia-lyase, AdoCbl/substrate-free | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Ethanolamine ammonia-lyase heavy chain, ... | | Authors: | Shibata, N. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Direct Participation of a Peripheral Side Chain of a Corrin Ring in Coenzyme B12Catalysis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YSH
 
 | |
3A20
 
 | |
3A2Q
 
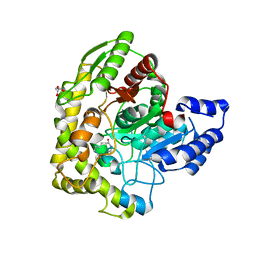 | | Structure of 6-aminohexanoate cyclic dimer hydrolase complexed with substrate | | Descriptor: | 6-AMINOHEXANOIC ACID, 6-aminohexanoate-cyclic-dimer hydrolase, GLYCEROL | | Authors: | Shibata, N. | | Deposit date: | 2009-05-26 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation
J.Biol.Chem., 285, 2010
|
|
3A2P
 
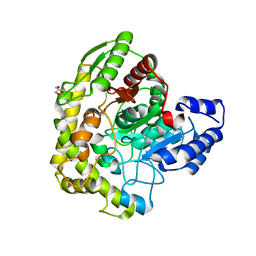 | | Structure of 6-aminohexanoate cyclic dimer hydrolase | | Descriptor: | 6-aminohexanoate-cyclic-dimer hydrolase, GLYCEROL | | Authors: | Shibata, N. | | Deposit date: | 2009-05-26 | | Release date: | 2009-11-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation
J.Biol.Chem., 285, 2010
|
|
3ANY
 
 | |
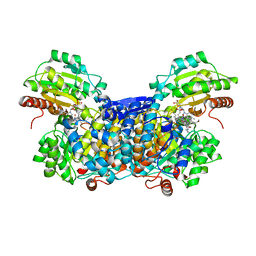
3AO0
 
 | |
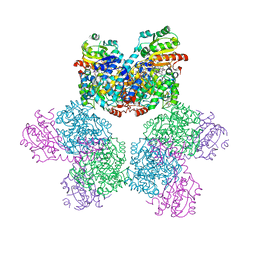
3ABQ
 
 | |
3ABR
 
 | |
3ABO
 
 | |
3ABS
 
 | |
1QVC
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SINGLE STRANDED DNA BINDING PROTEIN (SSB) FROM E.COLI | | Descriptor: | SINGLE STRANDED DNA BINDING PROTEIN MONOMER | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Shimamoto, N, Tsukihara, T, Yasuoka, N. | | Deposit date: | 1999-07-07 | | Release date: | 2000-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis.
J.Biochem.(Tokyo), 127, 2000
|
|
4UBQ
 
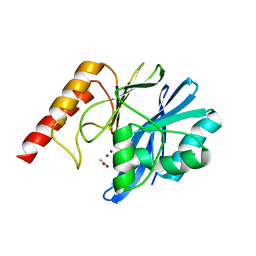 | | Crystal Structure of IMP-2 Metallo-beta-Lactamase from Acinetobacter spp. | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Yamaguchi, Y, Matsueda, S, Matsunaga, K, Takashio, N, Toma-Fukai, S, Yamagata, Y, Shibata, N, Wachino, J, Shibayama, K, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2.
Biol.Pharm.Bull., 38, 2015
|
|
7V5P
 
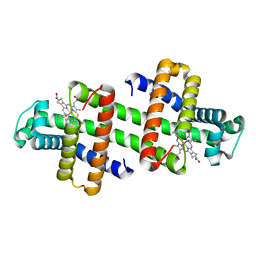 | | The dimeric structure of G80A/H81A myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xie, C, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Experimental and theoretical study on converting myoglobin into a stable domain-swapped dimer by utilizing a tight hydrogen bond network at the hinge region.
Rsc Adv, 11, 2021
|
|
6LS8
 
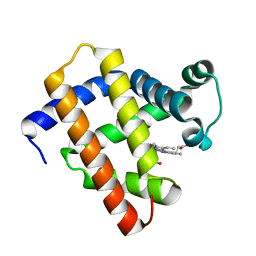 | | The monomeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTM
 
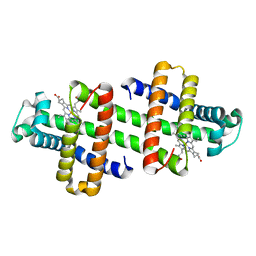 | | The dimeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTL
 
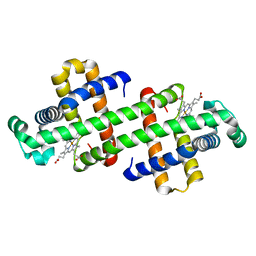 | | The dimeric structure of G80A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6L1V
 
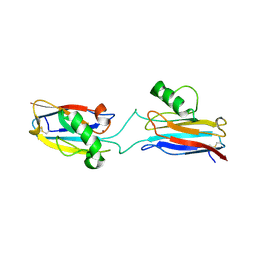 | | Domain-swapped Alcaligenes xylosoxidans azurin dimer | | Descriptor: | Azurin-1, COPPER (II) ION | | Authors: | Cahyono, R.N, Yamanaka, M, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3D domain swapping of azurin from Alcaligenes xylosoxidans.
Metallomics, 12, 2020
|
|
5GYR
 
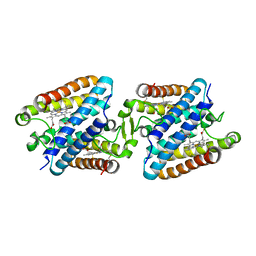 | | Tetrameric Allochromatium vinosum cytochrome c' | | Descriptor: | Cytochrome c', HEME C | | Authors: | Yamanaka, M, Hoshizumi, M, Nagao, S, Nakayama, R, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2016-09-23 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Formation and carbon monoxide-dependent dissociation of Allochromatium vinosum cytochrome c' oligomers using domain-swapped dimers
Protein Sci., 26, 2017
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
5AWE
 
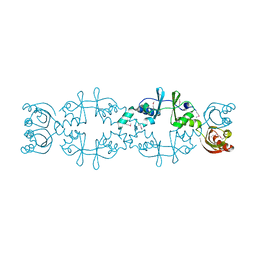 | | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains | | Descriptor: | Putative acetoin utilization protein, acetoin dehydrogenase | | Authors: | Nakabayashi, M, Shibata, N, Kanagawa, M, Nakagawa, N, Kuramitsu, S, Higuchi, Y. | | Deposit date: | 2015-07-03 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains.
Extremophiles, 20, 2016
|
|
5B7E
 
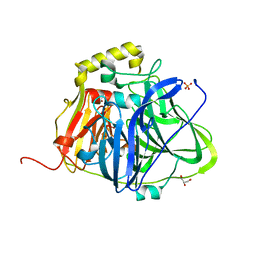 | | Structure of perdeuterated CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Akter, M, Higuchi, Y, Shibata, N. | | Deposit date: | 2016-06-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5B7F
 
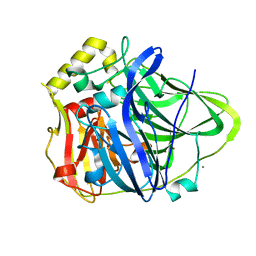 | | Structure of CueO - the signal peptide was truncated by HRV3C protease | | Descriptor: | 1,2-ETHANEDIOL, Blue copper oxidase CueO, CALCIUM ION, ... | | Authors: | Akter, M, Higuchi, Y, Shibata, N. | | Deposit date: | 2016-06-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1EGV
 
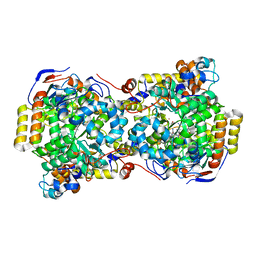 | | CRYSTAL STRUCTURE OF THE DIOL DEHYDRATASE-ADENINYLPENTYLCOBALAMIN COMPLEX FROM KLEBSELLA OXYTOCA UNDER THE ILLUMINATED CONDITION. | | Descriptor: | CO-(ADENIN-9-YL-PENTYL)-COBALAMIN, POTASSIUM ION, PROPANEDIOL DEHYDRATASE, ... | | Authors: | Masuda, J, Shibata, N, Toraya, T, Morimoto, Y, Yasuoka, N. | | Deposit date: | 2000-02-17 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How a protein generates a catalytic radical from coenzyme B(12): X-ray structure of a diol-dehydratase-adeninylpentylcobalamin complex.
Structure Fold.Des., 8, 2000
|
|
